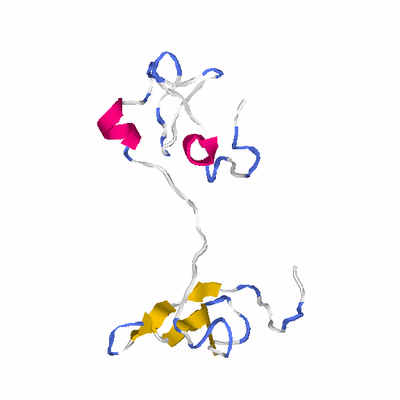
##########
# PROTEIN: Q2246_545_279.5wLII_11238_5 176 279 # Category_C1
# Length: 176
# Weight: 20265.33
# Similarity: UniProt_A3ER51, BLAST_A3ER51, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 54.55 M_06 1mo9_A 523 10 5e-08 18.000 96:116 (56-168:266-364)
# Evl 68.18 M_00 2vr5_A 718 6 2e-32 14.000 120:144 (8-150:509-629)
# Sid 43.75 M_00 2vr5_A 718 6 4e-21 20.000 77:79 (73-151:552-628)
# Cov 68.18 M_00 2vr5_A 718 6 2e-32 14.000 120:144 (8-150:509-629)
# LaL 26.70 M_02 2e8y_A 718 6 4e-10 20.000 47:48 (73-120:577-623)
# OvN 68.18 M_00 2vr5_A 718 6 2e-32 14.000 120:144 (8-150:509-629)
# OvC 56.82 M_08 3f8p_D 323 8 3e-07 12.000 100:122 (60-176:205-309)
PDB: 1mo9 PDBsum
HEADER OXIDOREDUCTASE 08-SEP-02 1MO9
TITLE NADPH DEPENDENT 2-KETOPROPYL COENZYME M
TITLE 2 OXIDOREDUCTASE/CARBOXYLASE COMPLEXED WITH 2-KETOPROPYL
TITLE 3 COENZYME M
SOURCE 2 ORGANISM_SCIENTIFIC: XANTHOBACTER AUTOTROPHICUS PY2;
SOURCE 3 ORGANISM_TAXID: 78245;
COMPND 6 EC: 1.6.4.-
KEYWDS NUCLEOTIDE BINDING MOTIFS, NUCLEOTIDE BINDING DOMAIN,
KEYWDS 2 DISULPHIDE BOND, OXIDOREDUCTASE
SCOP: c.3 FAD/NAD(P)-binding domain
SCOP: c.3.1 FAD/NAD(P)-binding domain
SCOP: c.3.1.5 FAD/NAD-linked reductases, N-terminal and central domains
SCOP: d.87 CO dehydrogenase flavoprotein C-domain-like
SCOP: d.87.1 FAD/NAD-linked reductases, dimerisation (C-terminal) domain
SCOP: d.87.1.1 FAD/NAD-linked reductases, dimerisation (C-terminal) domain
PDB: 2vr5 PDBsum
HEADER HYDROLASE 26-MAR-08 2VR5
TITLE CRYSTAL STRUCTURE OF TREX FROM SULFOLOBUS SOLFATARICUS IN
TITLE 2 COMPLEX WITH ACARBOSE INTERMEDIATE AND GLUCOSE
SOURCE 2 ORGANISM_SCIENTIFIC: SULFOLOBUS SOLFATARICUS;
SOURCE 3 ORGANISM_TAXID: 2287;
COMPND 5 EC: 3.2.1.-;
KEYWDS HYDROLASE, GLYCOSIDASE, GLYCOSYL HYDROLASE, GLYCOGEN
KEYWDS 2 DEBRACHING
SCOP: none
PDB: 2e8y PDBsum
HEADER HYDROLASE 24-JAN-07 2E8Y
TITLE CRYSTAL STRUCTURE OF PULLULANASE TYPE I FROM BACILLUS
TITLE 2 SUBTILIS STR. 168
SOURCE 2 ORGANISM_SCIENTIFIC: BACILLUS SUBTILIS;
SOURCE 3 ORGANISM_TAXID: 1423;
COMPND 5 EC: 3.2.1.41;
KEYWDS MULTIPLE DOMAIN, BETA-ALPHA-BARREL, ALPHA-AMYLASE-FAMILY,
KEYWDS 2 HYDROLASE
SCOP: none
PDB: 3f8p PDBsum
HEADER OXIDOREDUCTASE 13-NOV-08 3F8P
TITLE STRUCTURE OF SULFOLOBUS SOLFATARICUS TRXR-B3
SOURCE 2 ORGANISM_SCIENTIFIC: SULFOLOBUS SOLFATARICUS;
SOURCE 3 ORGANISM_TAXID: 2287;
COMPND 4 EC: 1.6.4.5;
KEYWDS REDOX PROTEIN, NUCLEOTIDE BINDING, FAD, FLAVOPROTEIN,
KEYWDS 2 OXIDOREDUCTASE
SCOP: none