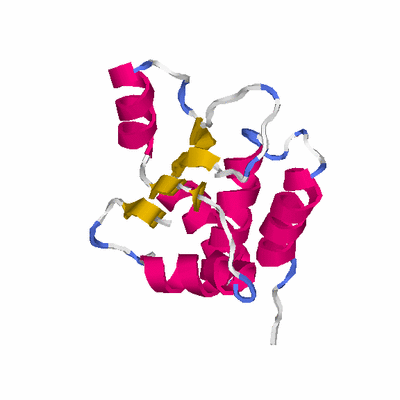
##########
# PROTEIN: Q2246_545_295.5wLII_11238_48 169 295 # Category_B
# Length: 169
# Weight: 19094.85
# Similarity: UniProt_B6AR29, BLAST_B6AR29, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 81.07 M_00 1xax_A 154 1 1e-33 27.000 137:140 (3-142:8-144)
# Evl 82.84 M_00 1xax_A 154 1 9e-36 26.000 140:147 (2-148:7-150)
# Sid 71.01 M_00 1oz9_A 150 1 1e-17 37.000 120:124 (23-146:29-150)
# Cov 86.98 M_02 1xm5_A 155 4 1e-27 24.000 147:155 (1-155:6-154)
# LaL 52.66 M_02 1xm5_A 155 4 1e-15 35.000 89:90 (32-121:36-124)
# OvN 85.80 M_01 1xm5_A 155 4 4e-34 25.000 145:148 (1-148:6-150)
# OvC 86.39 M_04 1tvi_A 172 1 2e-23 26.000 146:154 (3-156:24-169)
PDB: 1xax PDBsum
HEADER STRUCTURAL GENOMICS, UNKNOWN FUNCTION 26-AUG-04 1XAX
TITLE NMR STRUCTURE OF HI0004, A PUTATIVE ESSENTIAL GENE PRODUCT
TITLE 2 FROM HAEMOPHILUS INFLUENZAE
SOURCE 2 ORGANISM_SCIENTIFIC: HAEMOPHILUS INFLUENZAE;
SOURCE 3 ORGANISM_TAXID: 727;
KEYWDS HAEMOPHILUS INFLUENZAE, NMR, STRUCTURAL GENOMICS, MMP,
KEYWDS 2 HYDROLASE, PROTEIN STRUCTURE INITIATIVE, S2F, STRUCTURE 2
KEYWDS 3 FUNCTION PROJECT, UNKNOWN FUNCTION
SCOP: none
PDB: 1oz9 PDBsum
HEADER UNKNOWN FUNCTION 08-APR-03 1OZ9
TITLE CRYSTAL STRUCTURE OF AQ_1354, A HYPOTHETICAL PROTEIN FROM
TITLE 2 AQUIFEX AEOLICUS
SOURCE 2 ORGANISM_SCIENTIFIC: AQUIFEX AEOLICUS;
SOURCE 3 ORGANISM_TAXID: 63363;
KEYWDS MATRIX METALLOPROTEINASE TYPE FOLD, STRUCTURAL GENOMICS,
KEYWDS 2 BSGC STRUCTURE FUNDED BY NIH, PROTEIN STRUCTURE INITIATIVE,
KEYWDS 3 PSI, BERKELEY STRUCTURAL GENOMICS CENTER, UNKNOWN FUNCTION
SCOP: d.92 Zincin-like
SCOP: d.92.1 Metalloproteases ("zincins"), catalytic domain
SCOP: d.92.1.15 Predicted metal-dependent hydrolase
PDB: 1xm5 PDBsum
HEADER STRUCTURAL GENOMICS, UNKNOWN FUNCTION 01-OCT-04 1XM5
TITLE CRYSTAL STRUCTURE OF METAL-DEPENDENT HYDROLASE YBEY FROM E.
TITLE 2 COLI, PFAM UPF0054
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
SOURCE 3 ORGANISM_TAXID: 562;
KEYWDS STRUCTURAL GENOMICS, PROTEIN STRUCTURE INITIATIVE, NYSGXRC
KEYWDS 2 TARGET T842, PFAM FAMILY UPF0054, PSI, NEW YORK SGX
KEYWDS 3 RESEARCH CENTER FOR STRUCTURAL GENOMICS, UNKNOWN FUNCTION
SCOP: d.92 Zincin-like
SCOP: d.92.1 Metalloproteases ("zincins"), catalytic domain
SCOP: d.92.1.15 Predicted metal-dependent hydrolase
PDB: 1tvi PDBsum
HEADER STRUCTURAL GENOMICS, UNKNOWN FUNCTION 29-JUN-04 1TVI
TITLE SOLUTION STRUCTURE OF TM1509 FROM THERMOTOGA MARITIMA: VT1,
TITLE 2 A NESGC TARGET PROTEIN
SOURCE 2 ORGANISM_SCIENTIFIC: THERMOTOGA MARITIMA;
SOURCE 3 ORGANISM_TAXID: 2336;
KEYWDS ALPHA + BETA, MIXED 4-STRANDED BETA SHEET, FOUR HELIX
KEYWDS 2 BUNDLE, STRUCTURAL GENOMICS, PROTEIN STRUCTURE INITIATIVE,
KEYWDS 3 NESGC, PSI, NORTHEAST STRUCTURAL GENOMICS CONSORTIUM,
KEYWDS 4 UNKNOWN FUNCTION
SCOP: d.92 Zincin-like
SCOP: d.92.1 Metalloproteases ("zincins"), catalytic domain
SCOP: d.92.1.15 Predicted metal-dependent hydrolase