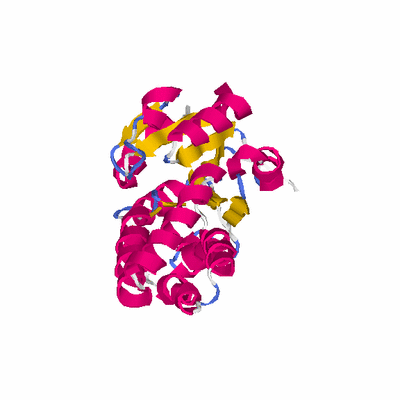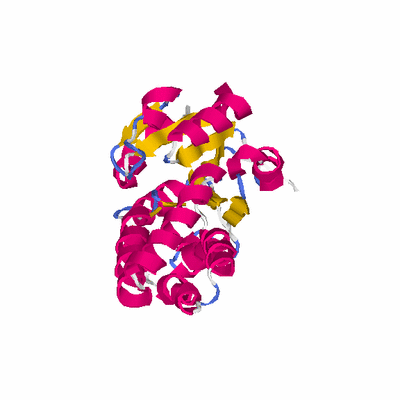##########
# PROTEIN: Q2246_545_309.5wLII_11238_93 238 309 # Category_A
# Length: 238
# Weight: 27519.36
# Similarity: UniProt_A3ERD8, BLAST_A3ERD8, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 90.76 M_00 z0qt 238 0 e-142 100.000 216:238 (1-238:1-238)
# Evl 90.76 M_00 z0qt 238 0 e-142 100.000 216:238 (1-238:1-238)
# Sid 90.76 M_00 z0qt 238 0 e-142 100.000 216:238 (1-238:1-238)
# Cov 92.02 M_06 2w4m_A 270 1 5e-31 12.000 219:257 (4-234:14-261)
# LaL 89.08 M_10 2p11_A 231 2 5e-30 45.000 212:214 (24-236:13-226)
# OvN 90.76 M_00 z0qt 238 0 e-142 100.000 216:238 (1-238:1-238)
# OvC 90.76 M_00 z0qt 238 0 e-142 100.000 216:238 (1-238:1-238)
PDB: 2p11 PDBsum
HEADER STRUCTURAL GENOMICS, UNKNOWN FUNCTION 01-MAR-07 2P11
TITLE CRYSTAL STRUCTURE OF A PUTATIVE HALOACID DELAHOGENASE-LIKE
TITLE 2 HYDROLASE (YP_553970.1) FROM BURKHOLDERIA XENOVORANS LB400
TITLE 3 AT 2.20 A RESOLUTION
SOURCE 2 ORGANISM_SCIENTIFIC: BURKHOLDERIA XENOVORANS LB400;
SOURCE 3 ORGANISM_TAXID: 266265;
KEYWDS YP_553970.1, HYPOTHETICAL PROTEIN, STRUCTURAL GENOMICS,
KEYWDS 2 JOINT CENTER FOR STRUCTURAL GENOMICS, JCSG, PROTEIN
KEYWDS 3 STRUCTURE INITIATIVE, PSI-2, UNKNOWN FUNCTION
SCOP: none
PDB: 2w4m PDBsum
HEADER HYDROLASE 28-NOV-08 2W4M
TITLE THE CRYSTAL STRUCTURE OF HUMAN N-ACETYLNEURAMINIC ACID
TITLE 2 PHOSPHATASE, NANP
SOURCE 2 ORGANISM_SCIENTIFIC: HOMO SAPIENS;
SOURCE 3 ORGANISM_COMMON: HUMAN;
SOURCE 4 ORGANISM_TAXID: 9606;
COMPND 6 EC: 3.1.3.29;
KEYWDS N-ACETYLNEURAMINATE, NEU5AC-9- PHOSPHATE, CARBOHYDRATE
KEYWDS 2 METABOLISM, N-ACETYLNEURAMINIC ACID PHOSPHATASE, NANP,
KEYWDS 3 HDHD4, HYDROLASE, MAGNESIUM, SIALIC ACID
SCOP: none