##########
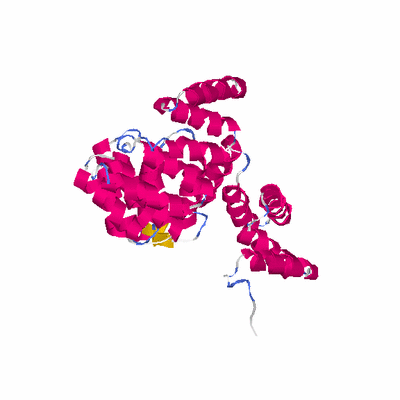
# PROTEIN: Q2246_545_312.5wLII_11238_104 556 312 # Category_C1
# Length: 556
# Weight: 60662.82
# Similarity: UniProt_A3ERE9, BLAST_A3ERE9, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 52.52 M_00 1w3b_A 388 2 9e-65 15.000 292:314 (2-311:81-376)
# Evl 48.74 M_00 2eyq_A 1151 2 2e-67 12.000 271:320 (26-306:444-753)
# Sid 21.76 M_00 3cv0_A 327 4 0.064 25.000 121:137 (249-370:147-283)
# Cov 66.01 M_04 2gw1_A 514 2 4e-27 9.000 367:478 (2-387:20-478)
# LaL 17.09 M_21 1wao_1 477 4 2e-12 11.000 95:95 (216-310:16-110)
# OvN 49.64 M_02 1gm5_A 780 1 7e-62 9.000 276:340 (1-306:204-518)
# OvC 64.75 M_04 2gw1_A 514 2 6e-23 12.000 360:468 (20-425:4-434)
PDB: 1w3b PDBsum
HEADER TRANSFERASE 14-JUL-04 1W3B
TITLE THE SUPERHELICAL TPR DOMAIN OF O-LINKED GLCNAC TRANSFERASE
TITLE 2 REVEALS STRUCTURAL SIMILARITIES TO IMPORTIN ALPHA.
SOURCE 2 ORGANISM_SCIENTIFIC: HOMO SAPIENS;
SOURCE 3 ORGANISM_COMMON: HUMAN;
SOURCE 4 ORGANISM_TAXID: 9606;
COMPND 7 EC: 2.4.1.-;
KEYWDS OGT, GLCNAC, NUCLEOPORIN, O-LINKED GLYCOSYLATION, TPR
KEYWDS 2 REPEAT, PROTEIN BINDING, SIGNAL TRANSDUCTION, TRANSFERASE
SCOP: a.118 alpha-alpha superhelix
SCOP: a.118.8 TPR-like
SCOP: a.118.8.1 Tetratricopeptide repeat (TPR)
PDB: 2eyq PDBsum
HEADER HYDROLASE 09-NOV-05 2EYQ
TITLE CRYSTAL STRUCTURE OF ESCHERICHIA COLI TRANSCRIPTION-REPAIR
TITLE 2 COUPLING FACTOR
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
SOURCE 3 ORGANISM_TAXID: 562;
KEYWDS MFD, SF2 ATPASE, HYDROLASE
SCOP: b.34 SH3-like barrel
SCOP: b.34.18 CarD-like
SCOP: b.34.18.1 CarD-like
SCOP: c.37 P-loop containing nucleoside triphosphate hydrolases
SCOP: c.37.1 P-loop containing nucleoside triphosphate hydrolases
SCOP: c.37.1.19 Tandem AAA-ATPase domain
SCOP: d.315 TRCF domain-like
SCOP: d.315.1 TRCF domain-like
SCOP: d.315.1.1 TRCF domain
PDB: 3cv0 PDBsum
HEADER TRANSPORT PROTEIN 17-APR-08 3CV0
TITLE STRUCTURE OF PEROXISOMAL TARGETING SIGNAL 1 (PTS1) BINDING
TITLE 2 DOMAIN OF TRYPANOSOMA BRUCEI PEROXIN 5 (TBPEX5)COMPLEXED
TITLE 3 TO T. BRUCEI PHOSPHOGLUCOISOMERASE (PGI) PTS1 PEPTIDE
SOURCE 2 ORGANISM_SCIENTIFIC: TRYPANOSOMA BRUCEI;
KEYWDS TPR MOTIFS, TPR PROTEIN, PEROXIN 5, PEX5, PTS1 BINDING
KEYWDS 2 DOMAIN, PROTEIN-PEPTIDE COMPLEX, RECEPTOR, TPR REPEAT,
KEYWDS 3 TRANSPORT PROTEIN
SCOP: none
PDB: 2gw1 PDBsum
HEADER PROTEIN TRANSPORT 03-MAY-06 2GW1
TITLE CRYSTAL STRUCTURE OF THE YEAST TOM70
SOURCE 2 ORGANISM_SCIENTIFIC: SACCHAROMYCES CEREVISIAE;
SOURCE 3 ORGANISM_COMMON: BAKER'S YEAST;
SOURCE 4 ORGANISM_TAXID: 4932;
KEYWDS TPR, PROTEIN TRANSPORT
SCOP: none
PDB: 1wao PDBsum
HEADER HYDROLASE 27-OCT-04 1WAO
TITLE PP5 STRUCTURE
SOURCE 2 ORGANISM_SCIENTIFIC: HOMO SAPIENS;
SOURCE 3 ORGANISM_COMMON: HUMAN;
SOURCE 4 ORGANISM_TAXID: 9606;
COMPND 6 EC: 3.1.3.16;
KEYWDS HYDROLASE, PHOSPHATASE, PROTEIN-PROTEIN INTERACTIONS, TPR,
KEYWDS 2 SUPER-HELIX, X-RAY STRUCTURE
SCOP: a.118 alpha-alpha superhelix
SCOP: a.118.8 TPR-like
SCOP: a.118.8.1 Tetratricopeptide repeat (TPR)
SCOP: d.159 Metallo-dependent phosphatases
SCOP: d.159.1 Metallo-dependent phosphatases
SCOP: d.159.1.3 Protein serine/threonine phosphatase
PDB: 1gm5 PDBsum
HEADER HELICASE 11-SEP-01 1GM5
TITLE STRUCTURE OF RECG BOUND TO THREE-WAY DNA JUNCTION
SOURCE 2 ORGANISM_SCIENTIFIC: THERMOTOGA MARITIMA;
SOURCE 3 ORGANISM_TAXID: 2336;
KEYWDS HELICASE, REPLICATION RESTART
SCOP: a.24 Four-helical up-and-down bundle
SCOP: a.24.21 RecG, N-terminal domain
SCOP: a.24.21.1 RecG, N-terminal domain
SCOP: b.40 OB-fold
SCOP: b.40.4 Nucleic acid-binding proteins
SCOP: b.40.4.9 RecG "wedge" domain
SCOP: c.37 P-loop containing nucleoside triphosphate hydrolases
SCOP: c.37.1 P-loop containing nucleoside triphosphate hydrolases
SCOP: c.37.1.19 Tandem AAA-ATPase domain