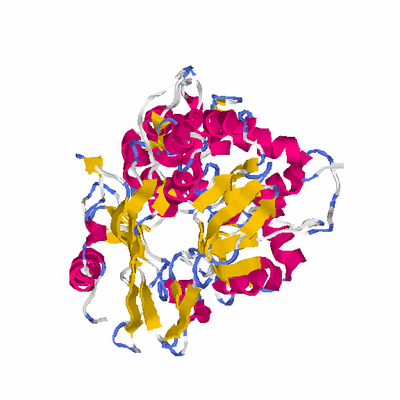
##########
# PROTEIN: Q2246_545_313.5wLII_11238_106 475 313 # Category_C1
# Length: 475
# Weight: 52790.84
# Similarity: UniProt_A3ERF1, BLAST_A3ERF1, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 87.37 M_25 2ivd_A 478 4 6e-24 15.000 415:470 (2-460:38-469)
# Evl 87.58 M_00 1o5w_C 534 4 2e-54 13.000 416:503 (4-474:45-492)
# Sid 5.05 M_30 2e1m_A 376 1 0.003 44.000 24:25 (2-25:66-90)
# Cov 97.89 M_17 2v1d_A 730 1 4e-07 11.000 465:544 (2-473:178-720)
# LaL 9.05 M_29 1sez_A 504 2 0.014 25.000 43:43 (3-45:36-78)
# OvN 86.53 M_08 2z5x_A 513 1 4e-53 12.000 411:491 (2-467:24-459)
# OvC 87.58 M_00 1o5w_C 534 4 2e-54 13.000 416:503 (4-474:45-492)
PDB: 2ivd PDBsum
HEADER OXIDOREDUCTASE 12-JUN-06 2IVD
TITLE STRUCTURE OF PROTOPORPHYRINOGEN OXIDASE FROM MYXOCOCCUS
TITLE 2 XANTHUS WITH ACIFLUORFEN
SOURCE 2 ORGANISM_SCIENTIFIC: MYXOCOCCUS XANTHUS;
SOURCE 3 ORGANISM_TAXID: 34;
COMPND 5 EC: 1.3.3.4;
KEYWDS PROTOPORPHYRINOGEN OXIDASE, PORPHYRIN BIOSYNTHESIS,
KEYWDS 2 CHLOROPHYLL BIOSYNTHESIS, OXIDOREDUCTASE, HAEM
KEYWDS 3 BIOSYNTHESIS, HEME BIOSYNTHESIS, FAD, PORPHYRIA,
KEYWDS 4 ACIFLUORFEN, FLAVOPROTEIN
SCOP: none
PDB: 1o5w PDBsum
HEADER OXIDOREDUCTASE 06-OCT-03 1O5W
TITLE THE STRUCTURE BASIS OF SPECIFIC RECOGNITIONS FOR SUBSTRATES
TITLE 2 AND INHIBITORS OF RAT MONOAMINE OXIDASE A
SOURCE 2 ORGANISM_SCIENTIFIC: RATTUS NORVEGICUS;
SOURCE 3 ORGANISM_COMMON: NORWAY RAT;
SOURCE 4 ORGANISM_TAXID: 10116;
COMPND 5 EC: 1.4.3.4;
KEYWDS OXIDOREDUCTASE
SCOP: c.3 FAD/NAD(P)-binding domain
SCOP: c.3.1 FAD/NAD(P)-binding domain
SCOP: c.3.1.2 FAD-linked reductases, N-terminal domain
SCOP: d.16 FAD-linked reductases, C-terminal domain
SCOP: d.16.1 FAD-linked reductases, C-terminal domain
SCOP: d.16.1.5 L-aminoacid/polyamine oxidase
PDB: 2e1m PDBsum
HEADER OXIDOREDUCTASE 26-OCT-06 2E1M
TITLE CRYSTAL STRUCTURE OF L-GLUTAMATE OXIDASE FROM STREPTOMYCES
TITLE 2 SP. X-119-6
SOURCE 2 ORGANISM_SCIENTIFIC: STREPTOMYCES SP. X-119-6;
SOURCE 3 ORGANISM_TAXID: 196112;
SOURCE 11 ORGANISM_SCIENTIFIC: STREPTOMYCES SP. X-119-6;
COMPND 5 EC: 1.4.3.11;
COMPND 11 EC: 1.4.3.11;
COMPND 17 EC: 1.4.3.11;
KEYWDS L-AMINO ACID OXIDASE, L-GLUTAMATE OXIDASE, FAD, L-GOX,
KEYWDS 2 FLAVOPROTEIN, 3D-STRUCTURE, OXIDOREDUCTASE
SCOP: none
PDB: 2v1d PDBsum
HEADER OXIDOREDUCTASE/REPRESSOR 23-MAY-07 2V1D
TITLE STRUCTURAL BASIS OF LSD1-COREST SELECTIVITY IN HISTONE H3
TITLE 2 RECOGNITION
SOURCE 2 ORGANISM_SCIENTIFIC: HOMO SAPIENS;
SOURCE 3 ORGANISM_COMMON: HUMAN;
SOURCE 4 ORGANISM_TAXID: 9606;
COMPND 7 EC: 1.-.-.-;
KEYWDS OXIDOREDUCTASE/REPRESSOR, HISTONE DEMETHYLASE, ALTERNATIVE
KEYWDS 2 SPLICING, AMINE OXIDASE, TRANSCRIPTION, OXIDOREDUCTASE,
KEYWDS 3 FAD, LSD1, FLAVIN, REPRESSOR, COILED COIL, TRANSCRIPTION
KEYWDS 4 REGULATION, OXIDOREDUCTASE/REPRESSOR COMPLEX CHROMATIN
KEYWDS 5 REMODELLING, HOST-VIRUS INTERACTION, NUCLEAR PROTEIN,
KEYWDS 6 PHOSPHORYLATION, CHROMATIN REGULATOR
SCOP: none
PDB: 1sez PDBsum
HEADER OXIDOREDUCTASE 19-FEB-04 1SEZ
TITLE CRYSTAL STRUCTURE OF PROTOPORPHYRINOGEN IX OXIDASE
SOURCE 2 ORGANISM_SCIENTIFIC: NICOTIANA TABACUM;
SOURCE 3 ORGANISM_COMMON: COMMON TOBACCO;
SOURCE 4 ORGANISM_TAXID: 4097;
COMPND 6 EC: 1.3.3.4;
KEYWDS FAD-BINDING, PARA-HYDROXY-BENZOATE-HYDROXYLASE FOLD (PHBH-
KEYWDS 2 FOLD), MONOTOPIC MEMBRANE-BINDING DOMAIN, OXIDOREDUCTASE
SCOP: c.3 FAD/NAD(P)-binding domain
SCOP: c.3.1 FAD/NAD(P)-binding domain
SCOP: c.3.1.2 FAD-linked reductases, N-terminal domain
SCOP: d.16 FAD-linked reductases, C-terminal domain
SCOP: d.16.1 FAD-linked reductases, C-terminal domain
SCOP: d.16.1.5 L-aminoacid/polyamine oxidase
PDB: 2z5x PDBsum
HEADER OXIDOREDUCTASE 20-JUL-07 2Z5X
TITLE CRYSTAL STRUCTURE OF HUMAN MONOAMINE OXIDASE A WITH HARMINE
SOURCE 2 ORGANISM_SCIENTIFIC: HOMO SAPIENS;
SOURCE 3 ORGANISM_COMMON: HUMAN;
SOURCE 4 ORGANISM_TAXID: 9606;
COMPND 6 EC: 1.4.3.4;
KEYWDS HUMAN MONOAMINE OXIDASE A, FAD, HARMINE,
KEYWDS 2 DIMETHYLDECYLPHOSPHINE OXIDE, SINGLE HELIX TRANS-MEMBRANE
KEYWDS 3 PROTEIN, ACETYLATION, CATECHOLAMINE METABOLISM,
KEYWDS 4 FLAVOPROTEIN, MITOCHONDRION, NEUROTRANSMITTER DEGRADATION,
KEYWDS 5 OXIDOREDUCTASE, POLYMORPHISM, TRANSMEMBRANE
SCOP: none