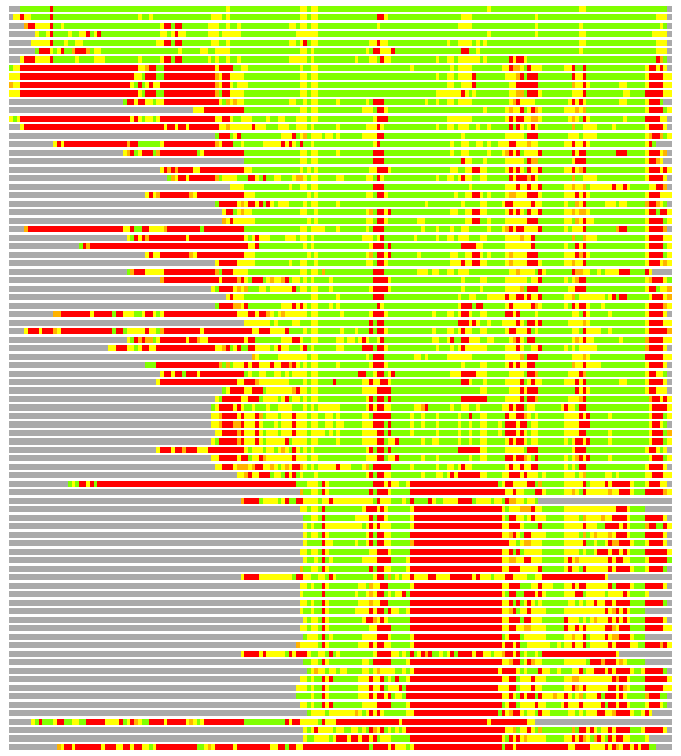
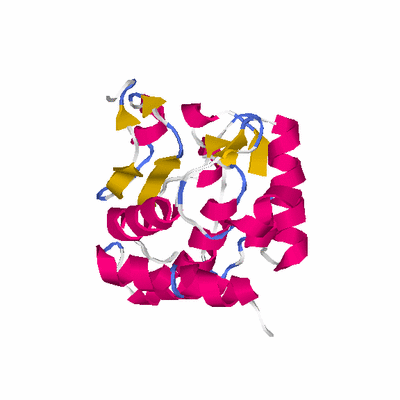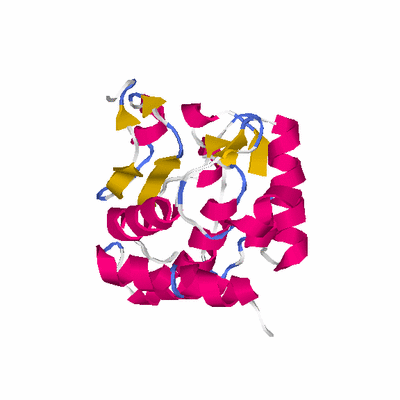##########
# PROTEIN: Q2246_545_331.5wLII_11276_77 189 331 # Category_C1
# Length: 189
# Weight: 21489.77
# Similarity: UniProt_B6ANY3, BLAST_B6ANY3, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 91.53 M_00 2yxe_A 215 2 5e-53 18.000 173:180 (3-177:1-179)
# Evl 91.53 M_00 2yxe_A 215 2 5e-53 18.000 173:180 (3-177:1-179)
# Sid 44.97 M_19 1xhe_B 123 2 3e-16 22.000 85:87 (102-187:3-88)
# Cov 97.88 M_01 1jg1_A 235 5 4e-45 16.000 185:198 (1-187:12-209)
# LaL 45.50 M_23 2gwr_A 238 1 7e-15 11.000 86:86 (103-188:6-91)
# OvN 89.95 M_01 1jg1_A 235 5 6e-52 15.000 170:183 (1-176:10-190)
# OvC 37.57 M_04 z0ct 240 0 8e-27 20.000 71:93 (104-189:7-98)
PDB: 2yxe PDBsum
HEADER TRANSFERASE 26-APR-07 2YXE
TITLE CRYSTAL STRUCTURE OF L-ISOASPARTYL PROTEIN CARBOXYL
TITLE 2 METHYLTRANFERASE
SOURCE 2 ORGANISM_SCIENTIFIC: METHANOCALDOCOCCUS JANNASCHII DSM
SOURCE 4 ORGANISM_TAXID: 243232;
COMPND 7 EC: 2.1.1.77;
KEYWDS ROSSMAN-TYPE FOLD, ALPHA/BETA/ALPHA SANDWICH STRUCTURE,
KEYWDS 2 STRUCTURAL GENOMICS, NPPSFA, NATIONAL PROJECT ON PROTEIN
KEYWDS 3 STRUCTURAL AND FUNCTIONAL ANALYSES, RIKEN STRUCTURAL
KEYWDS 4 GENOMICS/PROTEOMICS INITIATIVE, RSGI, TRANSFERASE
SCOP: none
PDB: 1xhe PDBsum
HEADER TRANSCRIPTION 18-SEP-04 1XHE
TITLE CRYSTAL STRUCTURE OF THE RECEIVER DOMAIN OF REDOX RESPONSE
TITLE 2 REGULATOR ARCA
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
SOURCE 3 ORGANISM_TAXID: 562;
KEYWDS TWO-COMPONENT SYSTEM; GENE REGULATION; TRANSCRIPTION
KEYWDS 2 FACTOR; ANOXIC REDOX CONTROL; DOUBLY WOUND FIVE-STRANDED
KEYWDS 3 BETA/ALPHA FOLD
SCOP: c.23 Flavodoxin-like
SCOP: c.23.1 CheY-like
SCOP: c.23.1.1 CheY-related
PDB: 1jg1 PDBsum
HEADER TRANSFERASE 22-JUN-01 1JG1
TITLE CRYSTAL STRUCTURE OF L-ISOASPARTYL (D-ASPARTYL) O-
TITLE 2 METHYLTRANSFERASE WITH S-ADENOSYL-L-HOMOCYSTEINE
SOURCE 2 ORGANISM_SCIENTIFIC: PYROCOCCUS FURIOSUS;
SOURCE 3 ORGANISM_TAXID: 2261;
COMPND 7 EC: 2.1.1.77;
KEYWDS ROSSMANN METHYLTRANSFERASE, PROTEIN REPAIR ISOMERIZATION
SCOP: c.66 S-adenosyl-L-methionine-dependent methyltransferases
SCOP: c.66.1 S-adenosyl-L-methionine-dependent methyltransferases
SCOP: c.66.1.7 Protein-L-isoaspartyl O-methyltransferase
PDB: 2gwr PDBsum
HEADER SIGNALING PROTEIN 05-MAY-06 2GWR
TITLE CRYSTAL STRUCTURE OF THE RESPONSE REGULATOR PROTEIN MTRA
TITLE 2 FROM MYCOBACTERIUM TUBERCULOSIS
SOURCE 2 ORGANISM_SCIENTIFIC: MYCOBACTERIUM TUBERCULOSIS;
SOURCE 3 ORGANISM_TAXID: 1773;
KEYWDS TWO-COMPONENT REGULATORY SYSTEM, TRANSCRIPTION REGULATION,
KEYWDS 2 PHOSPHORYLATION, DNA-BINDING, OMPR FAMILY, SIGNALING PROTEIN
SCOP: none
PDB: 1ys7 PDBsum
HEADER TRANSCRIPTION REGULATOR 07-FEB-05 1YS7
TITLE CRYSTAL STRUCTURE OF THE RESPONSE REGULATOR PROTEIN PRRA
TITLE 2 COMLEXED WITH MG2+
SOURCE 2 ORGANISM_SCIENTIFIC: MYCOBACTERIUM TUBERCULOSIS;
SOURCE 3 ORGANISM_TAXID: 1773;
KEYWDS RESPONSE REGULATOR, DNA BINDING DOMAIN, PHOSPHORYLATION,
KEYWDS 2 MYCOBACTERIUM TUBERCULOSIS STRUCTURAL PROTEOMICS PROJECT,
KEYWDS 3 XMTB, STRUCTURAL GENOMICS, TRANSCRIPTION REGULATOR
SCOP: a.4 DNA/RNA-binding 3-helical bundle
SCOP: a.4.6 C-terminal effector domain of the bipartite response regulators
SCOP: a.4.6.1 PhoB-like
SCOP: c.23 Flavodoxin-like
SCOP: c.23.1 CheY-like
SCOP: c.23.1.1 CheY-related