##########
# PROTEIN: Q2246_545_332.5wLII_11276_84 400 332 # Category_C1
# Length: 400
# Weight: 44231.18
# Similarity: UniProt_B6ANX6, BLAST_B6ANX6, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 89.00 M_00 2i09_B 403 2 4e-56 16.000 356:413 (2-392:5-401)
# Evl 85.00 M_00 z0l7 513 0 4e-78 10.000 340:557 (1-390:1-510)
# Sid 26.75 M_38 2i09_B 403 2 6e-05 26.000 107:116 (276-388:288-397)
# Cov 95.25 M_05 2ify_A 508 1 4e-49 10.000 381:516 (2-391:2-508)
# LaL 23.75 M_19 1y7a_A 449 2 5e-07 14.000 95:97 (235-330:275-370)
# OvN 85.00 M_00 z0l7 513 0 4e-78 10.000 340:557 (1-390:1-510)
# OvC 71.75 M_00 z00r 515 0 3e-48 15.000 287:372 (62-400:190-513)
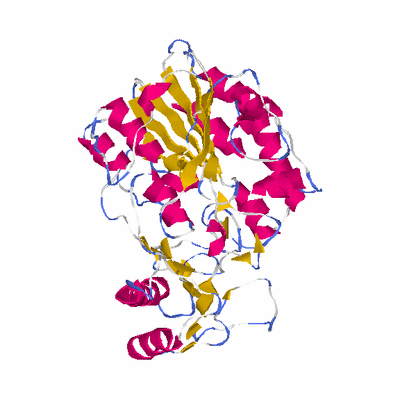
PDB: 2i09 PDBsum
HEADER ISOMERASE 10-AUG-06 2I09
TITLE CRYSTAL STRUCTURE OF PUTATIVE PHOSPHOPENTOMUTASE FROM
TITLE 2 STREPTOCOCCUS MUTANS
SOURCE 2 ORGANISM_SCIENTIFIC: STREPTOCOCCUS MUTANS;
SOURCE 3 ORGANISM_TAXID: 1309;
COMPND 5 EC: 5.4.2.7;
KEYWDS STRUCTURAL GENOMICS, TARGET T1865, NYSGXRC,
KEYWDS 2 PHOSPHOPENTOMUTASE, PSI, PROTEIN STRUCTURE INITIATIVE, NEW
KEYWDS 3 YORK SGX RESEARCH CENTER FOR STRUCTURAL GENOMICS, ISOMERASE
SCOP: c.76 Alkaline phosphatase-like
SCOP: c.76.1 Alkaline phosphatase-like
SCOP: c.76.1.5 DeoB catalytic domain-like
SCOP: d.327 DeoB insert domain-like
SCOP: d.327.1 DeoB insert domain-like
SCOP: d.327.1.1 DeoB insert domain-like
PDB: 1o98 PDBsum
HEADER ISOMERASE 11-DEC-02 1O98
TITLE 1.4A CRYSTAL STRUCTURE OF PHOSPHOGLYCERATE MUTASE FROM
TITLE 2 BACILLUS STEAROTHERMOPHILUS COMPLEXED WITH
TITLE 3 2-PHOSPHOGLYCERATE
SOURCE 2 ORGANISM_SCIENTIFIC: BACILLUS STEAROTHERMOPHILUS;
SOURCE 3 ORGANISM_TAXID: 1422;
COMPND 6 EC: 5.4.2.1;
KEYWDS ISOMERASE, ALPHA/BETA-TYPE STRUCTURE
SCOP: c.105 2,3-Bisphosphoglycerate-independent phosphoglycerate mutase, substrate-binding domain
SCOP: c.105.1 2,3-Bisphosphoglycerate-independent phosphoglycerate mutase, substrate-binding domain
SCOP: c.105.1.1 2,3-Bisphosphoglycerate-independent phosphoglycerate mutase, substrate-binding domain
SCOP: c.76 Alkaline phosphatase-like
SCOP: c.76.1 Alkaline phosphatase-like
SCOP: c.76.1.3 2,3-Bisphosphoglycerate-independent phosphoglycerate mutase, catalytic domain
PDB: 2ify PDBsum
HEADER ISOMERASE 21-SEP-06 2IFY
TITLE STRUCTURE OF BACILLUS ANTHRACIS COFACTOR-INDEPENDENT
TITLE 2 PHOSPHOGLUCERATE MUTASE
SOURCE 2 ORGANISM_SCIENTIFIC: BACILLUS ANTHRACIS;
SOURCE 3 ORGANISM_TAXID: 1392;
COMPND 6 EC: 5.4.2.1;
KEYWDS CATALYSIS, GLYGOLYSIS, GRAM-POSOTIVE BACTERIA, SPORES,
KEYWDS 2 CATALYTIC MECHANISM, PHOSPHOGLYCERATE, BACILLUS ANTHRACIS,
KEYWDS 3 ISOMERASE
SCOP: none
PDB: 1y7a PDBsum
HEADER HYDROLASE 08-DEC-04 1Y7A
TITLE STRUCTURE OF D153H/K328W E. COLI ALKALINE PHOSPHATASE IN
TITLE 2 PRESENCE OF COBALT AT 1.77 A RESOLUTION
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
SOURCE 3 ORGANISM_TAXID: 562;
COMPND 5 EC: 3.1.3.1;
KEYWDS METAL SPECIFICITY, X-RAY CRYSTALLOGRAPHY, HIGH-SPIN/LOW-
KEYWDS 2 SPIN CONFIGURATIONS, HYDROLASE
SCOP: none