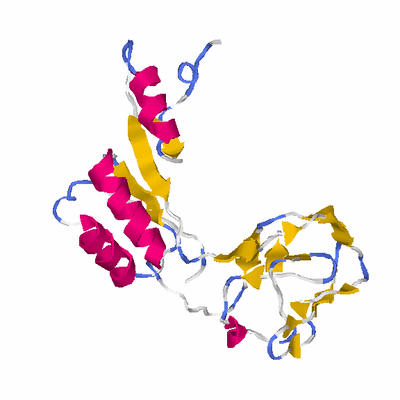
##########
# PROTEIN: Q2246_545_333.5wLII_11276_87 188 333 # Category_C1
# Length: 188
# Weight: 21368.46
# Similarity: UniProt_B6ANX3, BLAST_B6ANX3, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 97.87 M_00 1t5a_A 567 4 1e-59 15.000 184:214 (4-188:99-311)
# Evl 97.87 M_00 1t5a_A 567 4 1e-59 15.000 184:214 (4-188:99-311)
# Sid 71.28 M_16 2f7w_A 177 12 5e-28 21.000 134:155 (28-171:10-154)
# Cov 98.40 M_05 1e0u_A 470 4 4e-50 16.000 185:206 (3-188:21-225)
# LaL 76.60 M_25 2is8_B 164 3 1e-25 17.000 144:158 (28-177:8-159)
# OvN 98.40 M_05 1e0u_A 470 4 4e-50 16.000 185:206 (3-188:21-225)
# OvC 97.87 M_00 1t5a_A 567 4 1e-59 15.000 184:214 (4-188:99-311)
PDB: 1t5a PDBsum
HEADER TRANSFERASE 03-MAY-04 1T5A
TITLE HUMAN PYRUVATE KINASE M2
SOURCE 2 ORGANISM_SCIENTIFIC: HOMO SAPIENS;
SOURCE 3 ORGANISM_COMMON: HUMAN;
SOURCE 4 ORGANISM_TAXID: 9606;
COMPND 4 EC: 2.7.1.40;
KEYWDS ALPHA8-BETA8 BARREL, BETA SHEETS, ALPHA HELICES, TRANSFERASE
SCOP: none
PDB: 2f7w PDBsum
HEADER STRUCTURAL GENOMICS, UNKNOWN FUNCTION 01-DEC-05 2F7W
TITLE CRYSTAL STRUCTURE OF MOLYBDENUM COFACTOR BIOSYNTHESIS
TITLE 2 PROTEIN MOG FROM SHEWANELLA ONEIDENSIS
SOURCE 2 ORGANISM_SCIENTIFIC: SHEWANELLA ONEIDENSIS;
SOURCE 3 ORGANISM_TAXID: 70863;
KEYWDS STRUCTURAL GENOMICS MOLYBDENUM COFACTOR BIOSYNTHESIS, PSI,
KEYWDS 2 PROTEIN STRUCTURE INITIATIVE, MIDWEST CENTER FOR STRUCTURAL
KEYWDS 3 GENOMICS, MCSG, STRUCTURAL GENOMICS, UNKNOWN FUNCTION
SCOP: c.57 Molybdenum cofactor biosynthesis proteins
SCOP: c.57.1 Molybdenum cofactor biosynthesis proteins
SCOP: c.57.1.1 MogA-like
PDB: 1e0u PDBsum
HEADER PHOSPHOTRANSFERASE 10-APR-00 1E0U
TITLE STRUCTURE R271L MUTANT OF E. COLI PYRUVATE KINASE
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
SOURCE 3 ORGANISM_TAXID: 562;
COMPND 5 EC: 2.7.1.40;
KEYWDS PHOSPHOTRANSFERASE, GLYCOLYSIS, ALLOSTERY
SCOP: b.58 PK beta-barrel domain-like
SCOP: b.58.1 PK beta-barrel domain-like
SCOP: b.58.1.1 Pyruvate kinase beta-barrel domain
SCOP: c.1 TIM beta/alpha-barrel
SCOP: c.1.12 Phosphoenolpyruvate/pyruvate domain
SCOP: c.1.12.1 Pyruvate kinase
SCOP: c.49 Pyruvate kinase C-terminal domain-like
SCOP: c.49.1 PK C-terminal domain-like
SCOP: c.49.1.1 Pyruvate kinase, C-terminal domain
PDB: 2is8 PDBsum
HEADER STRUCTURAL PROTEIN 16-OCT-06 2IS8
TITLE CRYSTAL STRUCTURE OF THE MOLYBDOPTERIN BIOSYNTHESIS ENZYME
TITLE 2 MOAB (TTHA0341) FROM THERMUS THEROMOPHILUS HB8
SOURCE 2 ORGANISM_SCIENTIFIC: THERMUS THERMOPHILUS HB8;
SOURCE 3 ORGANISM_TAXID: 300852;
KEYWDS MOAB, GLOBULAR ALPHA/BETA FOLD, BIOSYNTHESIS ENZYME,
KEYWDS 2 STRUCTURAL GENOMICS, NPPSFA, NATIONAL PROJECT ON PROTEIN
KEYWDS 3 STRUCTURAL AND FUNCTIONAL ANALYSES, RIKEN STRUCTURAL
KEYWDS 4 GENOMICS/PROTEOMICS INITIATIVE, RSGI, STRUCTURAL PROTEIN
SCOP: none