##########
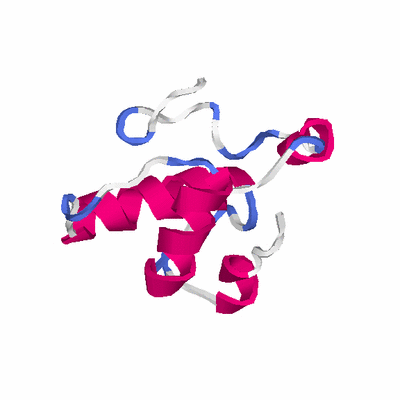
# PROTEIN: Q2246_545_337.5wLII_11276_107 92 337 # Category_B
# Length: 92
# Weight: 9696.91
# Similarity: UniProt_A3EV65, BLAST_A3EV65, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 83.70 M_02 z030 283 0 2e-07 26.000 77:102 (14-92:174-274)
# Evl 70.65 M_00 3bkh_A 268 2 4e-18 29.000 65:68 (24-90:8-73)
# Sid 43.48 M_02 z030 283 0 0.005 40.000 40:42 (31-71:210-251)
# Cov 95.65 M_02 z030 283 0 6e-07 28.000 88:107 (3-92:169-274)
# LaL 40.22 M_00 1lbu_A 213 1 0.019 40.000 37:37 (50-86:33-69)
# OvN 95.65 M_02 z030 283 0 6e-07 28.000 88:107 (3-92:169-274)
# OvC 65.22 M_02 z030 283 0 8e-09 34.000 60:63 (32-92:212-274)
PDB: 2bgx PDBsum
HEADER HYDROLASE 06-JAN-05 2BGX
TITLE CRYSTAL STRUCTURE OF NATIVE AMID AT 1.8 ANGSTROM
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
SOURCE 3 ORGANISM_TAXID: 562;
COMPND 4 EC: 3.5.1.28
KEYWDS ZINC AMIDASE, E. COLI, PGRP, T7 LYSOZYME, AMPD, N-
KEYWDS 2 ACETYLMUROAMYL-L-ALANINE AMIDASE, HYDROLASE
SCOP: a.20 PGBD-like
SCOP: a.20.1 PGBD-like
SCOP: a.20.1.1 Peptidoglycan binding domain, PGBD
SCOP: d.118 N-acetylmuramoyl-L-alanine amidase-like
SCOP: d.118.1 N-acetylmuramoyl-L-alanine amidase-like
SCOP: d.118.1.1 N-acetylmuramoyl-L-alanine amidase-like
PDB: 3bkh PDBsum
HEADER HYDROLASE 06-DEC-07 3BKH
TITLE CRYSTAL STRUCTURE OF THE BACTERIOPHAGE PHIKZ LYTIC
TITLE 2 TRANSGLYCOSYLASE, GP144
SOURCE 2 ORGANISM_SCIENTIFIC: PSEUDOMONAS PHAGE PHIKZ;
SOURCE 3 ORGANISM_TAXID: 169683;
KEYWDS TRANSGLYCOSYLASE, BACTERIOPHAGE, PHIKZ, ENDOLYSIN,
KEYWDS 2 PEPTIDOGLYCAN, CELL WALL DEGRADATION, LYSOZYME, HYDROLASE
SCOP: none
PDB: 1lbu PDBsum
HEADER HYDROLASE 16-MAR-96 1LBU
TITLE HYDROLASE METALLO (ZN) DD-PEPTIDASE
SOURCE 2 ORGANISM_SCIENTIFIC: STREPTOMYCES ALBUS G;
SOURCE 3 ORGANISM_TAXID: 1962;
COMPND 4 EC: 3.4.17.8
KEYWDS HYDROLASE, NUCLEAR RECEPTOR, CARBOXYPEPTIDASE
SCOP: a.20 PGBD-like
SCOP: a.20.1 PGBD-like
SCOP: a.20.1.1 Peptidoglycan binding domain, PGBD
SCOP: d.65 Hedgehog/DD-peptidase
SCOP: d.65.1 Hedgehog/DD-peptidase
SCOP: d.65.1.1 Muramoyl-pentapeptide carboxypeptidase