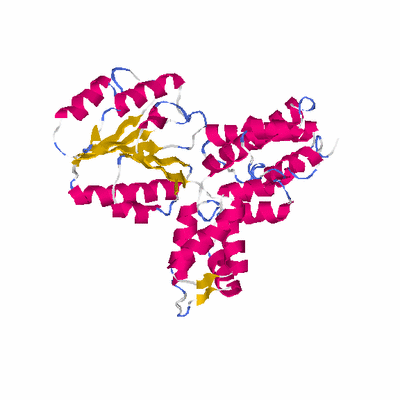
##########
# PROTEIN: Q2246_545_368.5wLII_11277_50 348 368 # Category_C1
# Length: 348
# Weight: 39210.24
# Similarity: UniProt_B6ALQ8, BLAST_B6ALQ8, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 95.40 M_03 1jr3_D 343 5 1e-41 16.000 332:343 (2-341:4-339)
# Evl 91.67 M_00 1jr3_A 373 9 8e-56 10.000 319:360 (5-341:27-368)
# Sid 91.67 M_05 z0me 344 0 2e-44 16.000 319:342 (6-342:10-344)
# Cov 95.40 M_03 1jr3_D 343 5 1e-41 16.000 332:343 (2-341:4-339)
# LaL 33.05 M_16 2r9g_L 204 16 2e-21 8.000 115:117 (207-321:11-127)
# OvN 93.39 M_00 z041 344 0 7e-54 13.000 325:333 (2-331:4-331)
# OvC 35.34 M_14 3bge_A 201 2 2e-24 8.000 123:145 (205-346:2-129)
PDB: 1jr3 PDBsum
HEADER TRANSFERASE 10-AUG-01 1JR3
TITLE CRYSTAL STRUCTURE OF THE PROCESSIVITY CLAMP LOADER GAMMA
TITLE 2 COMPLEX OF E. COLI DNA POLYMERASE III
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
SOURCE 3 ORGANISM_TAXID: 562;
SOURCE 7 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
COMPND 4 EC: 2.7.7.7;
COMPND 9 EC: 2.7.7.7;
COMPND 14 EC: 2.7.7.7;
KEYWDS DNA POLYMERASE, PROCESSIVITY, PROCESSIVITY CLAMP, CLAMP
KEYWDS 2 LOADER, AAA+ ATPASE, TRANSFERASE
SCOP: a.80 DNA polymerase III clamp loader subunits, C-terminal domain
SCOP: a.80.1 DNA polymerase III clamp loader subunits, C-terminal domain
SCOP: a.80.1.1 DNA polymerase III clamp loader subunits, C-terminal domain
SCOP: c.37 P-loop containing nucleoside triphosphate hydrolases
SCOP: c.37.1 P-loop containing nucleoside triphosphate hydrolases
SCOP: c.37.1.20 Extended AAA-ATPase domain
PDB: 2r9g PDBsum
HEADER HYDROLASE 12-SEP-07 2R9G
TITLE CRYSTAL STRUCTURE OF THE C-TERMINAL FRAGMENT OF AAA ATPASE
TITLE 2 FROM ENTEROCOCCUS FAECIUM
SOURCE 2 ORGANISM_SCIENTIFIC: ENTEROCOCCUS FAECIUM DO;
SOURCE 3 ORGANISM_TAXID: 333849;
KEYWDS STRUCTURAL GENOMICS, ATPASE, PSI-2, PROTEIN STRUCTURE
KEYWDS 2 INITIATIVE, NEW YORK SGX RESEARCH CENTER FOR STRUCTURAL
KEYWDS 3 GENOMICS, NYSGXRC, ATP-BINDING, NUCLEOTIDE-BINDING,
KEYWDS 4 HYDROLASE
SCOP: none
PDB: 3bge PDBsum
HEADER NUCLEOTIDE BINDING PROTEIN 26-NOV-07 3BGE
TITLE CRYSTAL STRUCTURE OF THE C-TERMINAL FRAGMENT OF AAA+ATPASE
TITLE 2 FROM HAEMOPHILUS INFLUENZAE
SOURCE 2 ORGANISM_SCIENTIFIC: HAEMOPHILUS INFLUENZAE 86-028NP;
SOURCE 3 ORGANISM_TAXID: 281310;
KEYWDS STRUCTURAL GENOMICS, PREDICTED AAA+ATPASE C-TERMINAL
KEYWDS 2 FRAGMENT, PSI-2, PROTEIN STRUCTURE INITIATIVE, NEW YORK SGX
KEYWDS 3 RESEARCH CENTER FOR STRUCTURAL GENOMICS, NYSGXRC, ATP-
KEYWDS 4 BINDING, HELICASE, NUCLEOTIDE-BINDING, NUCLEOTIDE BINDING
KEYWDS 5 PROTEIN
SCOP: none