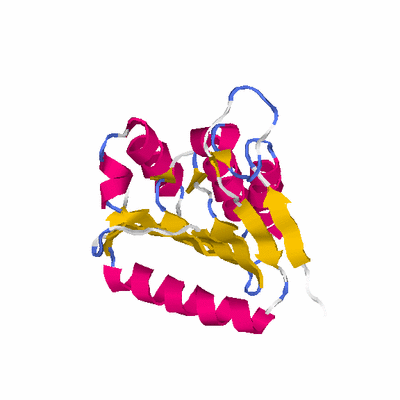
##########
# PROTEIN: Q2246_545_388.5wLII_11277_106 253 388 # Category_C1
# Length: 253
# Weight: 29802.14
# Similarity: UniProt_B6ALW2, BLAST_B6ALW2, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 58.89 M_00 1byr_A 155 2 1e-29 24.000 149:166 (65-230:4-152)
# Evl 59.29 M_00 1byr_A 155 2 8e-31 23.000 150:167 (64-230:3-152)
# Sid 36.76 M_00 1byr_A 155 2 8e-10 26.000 93:95 (80-174:19-111)
# Cov 79.84 M_01 2ze4_A 509 1 4e-05 12.000 202:241 (18-232:4-231)
# LaL 37.55 M_00 1byr_A 155 2 5e-10 25.000 95:97 (80-176:19-113)
# OvN 77.47 M_01 2ze4_A 509 1 2e-06 12.000 196:232 (16-224:4-222)
# OvC 72.33 M_02 2o8r_A 705 2 1e-06 14.000 183:201 (55-247:501-691)
PDB: 1byr PDBsum
HEADER ENDONUCLEASE 19-OCT-98 1BYR
TITLE CRYSTAL STRUCTURE OF A PHOSPHOLIPASE D FAMILY MEMBER, NUC
TITLE 2 FROM SALMONELLA TYPHIMURIUM
SOURCE 2 ORGANISM_SCIENTIFIC: SALMONELLA TYPHIMURIUM;
SOURCE 3 ORGANISM_TAXID: 602;
KEYWDS ENDONUCLEASE, PHOSPHODIESTERASE,
SCOP: d.136 Phospholipase D/nuclease
SCOP: d.136.1 Phospholipase D/nuclease
SCOP: d.136.1.1 Nuclease
PDB: 2ze4 PDBsum
HEADER HYDROLASE 05-DEC-07 2ZE4
TITLE CRYSTAL STRUCTURE OF PHOSPHOLIPASE D FROM STREPTOMYCES
TITLE 2 ANTIBIOTICUS
SOURCE 2 ORGANISM_SCIENTIFIC: STREPTOMYCES ANTIBIOTICUS;
SOURCE 3 ORGANISM_TAXID: 1890;
COMPND 5 EC: 3.1.4.4;
KEYWDS ALPHA-BETA-BETA-ALPHA-SANDWICH, HYDROLASE, LIPID
KEYWDS 2 DEGRADATION, SECRETED
SCOP: none
PDB: 2o8r PDBsum
HEADER TRANSFERASE 12-DEC-06 2O8R
TITLE CRYSTAL STRUCTURE OF POLYPHOSPHATE KINASE FROM
TITLE 2 PORPHYROMONAS GINGIVALIS
SOURCE 2 ORGANISM_SCIENTIFIC: PORPHYROMONAS GINGIVALIS;
SOURCE 3 ORGANISM_TAXID: 837;
COMPND 4 EC: 2.7.4.1;
KEYWDS KINASE, STRUCTURAL GENOMICS, PROTEIN STRUCTURE INITIATIVE,
KEYWDS 2 PSI, NYSGRC, NEW YORK STRUCTURAL GENOMICS RESEARCH
KEYWDS 3 CONSORTIUM, NEW YORK SGX RESEARCH CENTER FOR STRUCTURAL
KEYWDS 4 GENOMICS, NYSGXRC, TRANSFERASE
SCOP: a.7 Spectrin repeat-like
SCOP: a.7.15 PPK N-terminal domain-like
SCOP: a.7.15.1 PPK N-terminal domain-like
SCOP: d.322 PHP14-like
SCOP: d.322.1 PHP14-like
SCOP: d.322.1.2 PPK middle domain-like
SCOP: d.136 Phospholipase D/nuclease
SCOP: d.136.1 Phospholipase D/nuclease
SCOP: d.136.1.4 Polyphosphate kinase C-terminal domain