##########
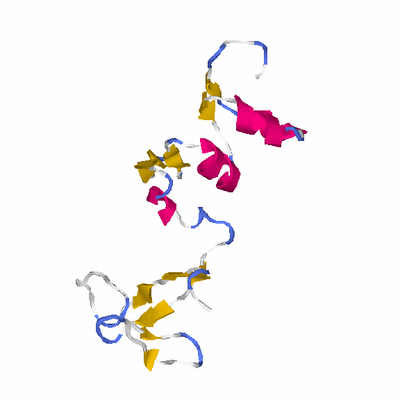
# PROTEIN: Q2246_545_393.5wLII_11277_121 126 393 # Category_B
# Length: 126
# Weight: 14514.07
# Similarity: UniProt_B6ALX7, BLAST_B6ALX7, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 94.44 M_01 z0am 357 0 7e-41 21.000 119:140 (3-125:150-285)
# Evl 96.03 M_00 z09b 366 0 6e-42 18.000 121:140 (3-125:149-286)
# Sid 89.68 M_00 2d1p_B 119 3 0.057 24.000 113:120 (5-119:2-119)
# Cov 98.41 M_12 2d62_A 375 1 2e-35 19.000 124:142 (2-125:156-297)
# LaL 65.08 M_20 3fve_A 290 1 6e-07 9.000 82:82 (23-104:68-149)
# OvN 96.83 M_00 1m7x_B 617 4 1e-40 15.000 122:138 (1-126:418-551)
# OvC 96.83 M_00 1m7x_B 617 4 1e-40 15.000 122:138 (1-126:418-551)
PDB: 2awn PDBsum
HEADER TRANSPORT PROTEIN 01-SEP-05 2AWN
TITLE CRYSTAL STRUCTURE OF THE ADP-MG-BOUND E. COLI MALK
TITLE 2 (CRYSTALLIZED WITH ATP-MG)
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI K12;
SOURCE 3 ORGANISM_TAXID: 83333;
COMPND 5 EC: 3.6.3.19;
KEYWDS ATP-BINDING CASSETTE, TRANSPORT PROTEIN
SCOP: b.40 OB-fold
SCOP: b.40.6 MOP-like
SCOP: b.40.6.3 ABC-transporter additional domain
SCOP: c.37 P-loop containing nucleoside triphosphate hydrolases
SCOP: c.37.1 P-loop containing nucleoside triphosphate hydrolases
SCOP: c.37.1.12 ABC transporter ATPase domain-like
PDB: 2d1p PDBsum
HEADER TRANSLATION 30-AUG-05 2D1P
TITLE CRYSTAL STRUCTURE OF HETEROHEXAMERIC TUSBCD PROTEINS, WHICH
TITLE 2 ARE CRUCIAL FOR THE TRNA MODIFICATION
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
SOURCE 3 ORGANISM_TAXID: 562;
SOURCE 11 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
KEYWDS TRNA MODIFICATION, SULFUR TRANSFER, STRUCTURAL GENOMICS,
KEYWDS 2 TRANSLATION
SCOP: c.114 DsrEFH-like
SCOP: c.114.1 DsrEFH-like
SCOP: c.114.1.1 DsrEF-like
SCOP: c.114 DsrEFH-like
SCOP: c.114.1 DsrEFH-like
SCOP: c.114.1.2 DsrH-like
PDB: 2d62 PDBsum
HEADER SUGAR BINDING PROTEIN 08-NOV-05 2D62
TITLE CRYSTAL STRUCTURE OF MULTIPLE SUGAR BINDING TRANSPORT ATP-
TITLE 2 BINDING PROTEIN
SOURCE 2 ORGANISM_SCIENTIFIC: PYROCOCCUS HORIKOSHII;
SOURCE 3 ORGANISM_TAXID: 53953;
KEYWDS ATP-BINDING, STRUCTURAL GEOMICS, STRUCTURAL GENOMICS,
KEYWDS 2 NPPSFA, NATIONAL PROJECT ON PROTEIN STRUCTURAL AND
KEYWDS 3 FUNCTIONAL ANALYSES, RIKEN STRUCTURAL GENOMICS/PROTEOMICS
KEYWDS 4 INITIATIVE, RSGI, SUGAR BINDING PROTEIN
SCOP: none
PDB: 3fve PDBsum
HEADER ISOMERASE 15-JAN-09 3FVE
TITLE CRYSTAL STRUCTURE OF DIAMINOPIMELATE EPIMERASE
TITLE 2 MYCOBACTERIUM TUBERCULOSIS DAPF
SOURCE 2 ORGANISM_SCIENTIFIC: MYCOBACTERIUM TUBERCULOSIS;
SOURCE 3 ORGANISM_TAXID: 1773;
COMPND 5 EC: 5.1.1.7;
KEYWDS ALPHA/BETA, AMINO-ACID BIOSYNTHESIS, CYTOPLASM, ISOMERASE,
KEYWDS 2 LYSINE BIOSYNTHESIS
SCOP: none
PDB: 1m7x PDBsum
HEADER TRANSFERASE 23-JUL-02 1M7X
TITLE THE X-RAY CRYSTALLOGRAPHIC STRUCTURE OF BRANCHING ENZYME
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
SOURCE 3 ORGANISM_TAXID: 562;
COMPND 6 EC: 2.4.1.18;
KEYWDS ALPHA/BETA BARREL, BETA SANDWICH, TRANSFERASE
SCOP: b.1 Immunoglobulin-like beta-sandwich
SCOP: b.1.18 E set domains
SCOP: b.1.18.2 E-set domains of sugar-utilizing enzymes
SCOP: b.71 Glycosyl hydrolase domain
SCOP: b.71.1 Glycosyl hydrolase domain
SCOP: b.71.1.1 alpha-Amylases, C-terminal beta-sheet domain
SCOP: c.1 TIM beta/alpha-barrel
SCOP: c.1.8 (Trans)glycosidases
SCOP: c.1.8.1 Amylase, catalytic domain