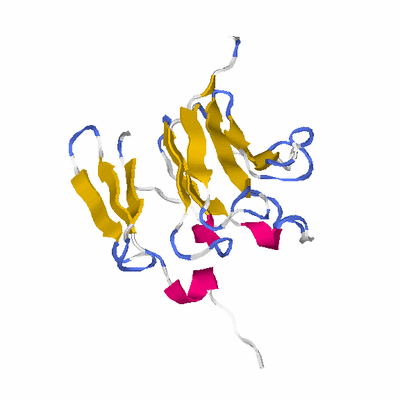
##########
# PROTEIN: Q2246_545_395.5wLII_11277_123 184 395 # Category_C1
# Length: 184
# Weight: 21150.16
# Similarity: UniProt_B6ALX9, BLAST_B6ALX9, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 73.91 M_00 3c99_A 432 2 2e-37 19.000 136:161 (4-162:280-428)
# Evl 90.76 M_00 1p3w_B 404 2 3e-58 7.000 167:193 (2-180:81-261)
# Sid 23.37 M_01 1g0d_A 695 1 0.064 27.000 43:43 (25-67:168-210)
# Cov 93.48 M_15 1vjo_A 393 1 3e-15 8.000 172:210 (2-184:96-294)
# LaL 36.41 M_11 2cot_A 77 1 9e-07 2.000 67:67 (113-179:10-76)
# OvN 90.76 M_00 1p3w_B 404 2 3e-58 7.000 167:193 (2-180:81-261)
# OvC 93.48 M_15 1vjo_A 393 1 3e-15 8.000 172:210 (2-184:96-294)
PDB: 3c99 PDBsum
HEADER TRANSCRIPTION REPRESSOR 15-FEB-08 3C99
TITLE STRUCTURAL BASIS OF HISTONE H4 RECOGNITION BY P55
SOURCE 2 ORGANISM_SCIENTIFIC: DROSOPHILA MELANOGASTER;
SOURCE 3 ORGANISM_COMMON: FRUIT FLY;
SOURCE 4 ORGANISM_TAXID: 7227;
KEYWDS WD40, HISTONE BINDING, CHROMATIN, EPIGENETICS, CHROMATIN
KEYWDS 2 REGULATOR, NUCLEUS, PHOSPHOPROTEIN, REPRESSOR,
KEYWDS 3 TRANSCRIPTION, TRANSCRIPTION REGULATION, WD REPEAT, NUCLEAR
KEYWDS 4 PROTEIN, TRANSCRIPTION REPRESSOR
SCOP: none
PDB: 1p3w PDBsum
HEADER LYASE 18-APR-03 1P3W
TITLE X-RAY CRYSTAL STRUCTURE OF E. COLI ISCS
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
SOURCE 3 ORGANISM_TAXID: 562;
COMPND 5 EC: 4.4.1.-;
KEYWDS IRON SULFUR CLUSTER, NIFS, CSDB, LYASE
SCOP: c.67 PLP-dependent transferases
SCOP: c.67.1 PLP-dependent transferases
SCOP: c.67.1.3 Cystathionine synthase-like
PDB: 1g0d PDBsum
HEADER TRANSFERASE 06-OCT-00 1G0D
TITLE CRYSTAL STRUCTURE OF RED SEA BREAM TRANSGLUTAMINASE
SOURCE 2 ORGANISM_SCIENTIFIC: PAGRUS MAJOR;
SOURCE 3 ORGANISM_COMMON: RED SEABREAM;
SOURCE 4 ORGANISM_TAXID: 143350;
COMPND 4 EC: 2.3.2.13;
KEYWDS TISSUE TRANSGLUTAMINASE,ACYLTRANSFERASE
SCOP: b.1 Immunoglobulin-like beta-sandwich
SCOP: b.1.18 E set domains
SCOP: b.1.18.9 Transglutaminase N-terminal domain
SCOP: b.1 Immunoglobulin-like beta-sandwich
SCOP: b.1.5 Transglutaminase, two C-terminal domains
SCOP: b.1.5.1 Transglutaminase, two C-terminal domains
SCOP: d.3 Cysteine proteinases
SCOP: d.3.1 Cysteine proteinases
SCOP: d.3.1.4 Transglutaminase core
PDB: 1vjo PDBsum
HEADER TRANSFERASE 16-MAR-04 1VJO
TITLE CRYSTAL STRUCTURE OF ALANINE--GLYOXYLATE AMINOTRANSFERASE
TITLE 2 (ALR1004) FROM NOSTOC SP. AT 1.70 A RESOLUTION
SOURCE 2 ORGANISM_SCIENTIFIC: NOSTOC SP. PCC 7120;
SOURCE 3 ORGANISM_TAXID: 103690;
COMPND 4 EC: 2.6.1.44;
KEYWDS 17130350, ALR1004, ALANINE--GLYOXYLATE AMINOTRANSFERASE,
KEYWDS 2 STRUCTURAL GENOMICS, JCSG, PSI, PROTEIN STRUCTURE
KEYWDS 3 INITIATIVE, JOINT CENTER FOR STRUCTURAL GENOMICS
SCOP: c.67 PLP-dependent transferases
SCOP: c.67.1 PLP-dependent transferases
SCOP: c.67.1.3 Cystathionine synthase-like
PDB: 2cot PDBsum
HEADER TRANSCRIPTION 18-MAY-05 2COT
TITLE SOLUTION STRUCTURE OF THE FIRST AND SECOND ZF-C2H2 DOMAIN
TITLE 2 OF ZINC FINGER PROTEIN 435
SOURCE 2 ORGANISM_SCIENTIFIC: HOMO SAPIENS;
SOURCE 3 ORGANISM_COMMON: HUMAN;
SOURCE 4 ORGANISM_TAXID: 9606;
KEYWDS ADK_LID DOMAIN, ZINC FINGER PROTEIN 435, ZINC FINGER AND
KEYWDS 2 SCAN DOMAIN CONTAINING PROTEIN 16, STRUCTURAL GENOMICS,
KEYWDS 3 NPPSFA, NATIONAL PROJECT ON PROTEIN STRUCTURAL AND
KEYWDS 4 FUNCTIONAL ANALYSES, RIKEN STRUCTURAL GENOMICS/PROTEOMICS
KEYWDS 5 INITIATIVE, RSGI, TRANSCRIPTION
SCOP: g.37 C2H2 and C2HC zinc fingers
SCOP: g.37.1 C2H2 and C2HC zinc fingers
SCOP: g.37.1.1 Classic zinc finger, C2H2