##########
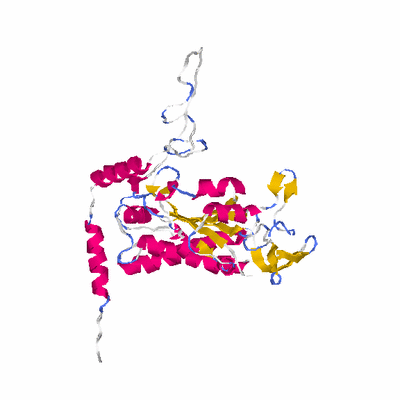
# PROTEIN: Q2246_545_411.5wLII_11277_158 600 411 # Category_C2
# Length: 600
# Weight: 67140.94
# Similarity: UniProt_B6AM13, BLAST_B6AM13, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 43.67 M_00 1p4d_B 330 3 3e-85 21.000 262:360 (1-347:2-330)
# Evl 43.67 M_00 1p4d_B 330 3 3e-85 21.000 262:360 (1-347:2-330)
# Sid 16.00 M_08 1ffh_A 294 1 0.090 32.000 96:116 (364-479:17-112)
# Cov 56.83 M_11 1qvr_A 854 3 5e-06 11.000 341:414 (200-570:290-687)
# LaL 7.83 M_38 z0md 347 0 0.034 19.000 47:47 (457-503:116-162)
# OvN 43.67 M_00 1p4d_B 330 3 3e-85 21.000 262:360 (1-347:2-330)
# OvC 23.83 M_08 2is6_A 680 8 2e-31 16.000 143:163 (444-600:6-154)
PDB: 1p4d PDBsum
HEADER HYDROLASE 22-APR-03 1P4D
TITLE F FACTOR TRAI RELAXASE DOMAIN
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
SOURCE 3 ORGANISM_TAXID: 562;
COMPND 7 EC: 3.6.1.-;
KEYWDS 5-STRAND ANTIPARALLEL BETA SHEET, ALPHA-BETA, 3 HISTIDINE
KEYWDS 2 MG(II) COORDINATION, HYDROLASE
SCOP: d.89 Origin of replication-binding domain, RBD-like
SCOP: d.89.1 Origin of replication-binding domain, RBD-like
SCOP: d.89.1.5 Relaxase domain
PDB: 1ffh PDBsum
HEADER RIBONUCLEOPROTEIN 30-DEC-96 1FFH
TITLE N AND GTPASE DOMAINS OF THE SIGNAL SEQUENCE RECOGNITION
TITLE 2 PROTEIN FFH FROM THERMUS AQUATICUS
SOURCE 2 ORGANISM_SCIENTIFIC: THERMUS AQUATICUS;
SOURCE 3 ORGANISM_TAXID: 271;
KEYWDS FFH, SRP, GTPASE, SIGNAL RECOGNITION PARTICLE,
KEYWDS 2 RIBONUCLEOPROTEIN
SCOP: a.24 Four-helical up-and-down bundle
SCOP: a.24.13 Domain of the SRP/SRP receptor G-proteins
SCOP: a.24.13.1 Domain of the SRP/SRP receptor G-proteins
SCOP: c.37 P-loop containing nucleoside triphosphate hydrolases
SCOP: c.37.1 P-loop containing nucleoside triphosphate hydrolases
SCOP: c.37.1.10 Nitrogenase iron protein-like
PDB: 1qvr PDBsum
HEADER CHAPERONE 28-AUG-03 1QVR
TITLE CRYSTAL STRUCTURE ANALYSIS OF CLPB
SOURCE 2 ORGANISM_SCIENTIFIC: THERMUS THERMOPHILUS;
SOURCE 3 ORGANISM_TAXID: 274;
KEYWDS COILED COIL, AAA ATPASE, CHAPERONE
SCOP: a.174 Double Clp-N motif
SCOP: a.174.1 Double Clp-N motif
SCOP: a.174.1.1 Double Clp-N motif
SCOP: c.37 P-loop containing nucleoside triphosphate hydrolases
SCOP: c.37.1 P-loop containing nucleoside triphosphate hydrolases
SCOP: c.37.1.20 Extended AAA-ATPase domain
PDB: 2ewv PDBsum
HEADER PROTEIN TRANSPORT 07-NOV-05 2EWV
TITLE CRYSTAL STRUCTURE OF THE PILUS RETRACTION MOTOR PILT AND
TITLE 2 BOUND ADP
SOURCE 2 ORGANISM_SCIENTIFIC: AQUIFEX AEOLICUS VF5;
SOURCE 3 ORGANISM_TAXID: 224324;
KEYWDS PILUS RETRACTION MOTOR, ATPASE, HEXAMERIC PILT, PROTEIN
KEYWDS 2 TRANSPORT
SCOP: none
PDB: 2is6 PDBsum
HEADER HYDROLASE/DNA 16-OCT-06 2IS6
TITLE CRYSTAL STRUCTURE OF UVRD-DNA-ADPMGF3 TERNARY COMPLEX
SOURCE 4 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
SOURCE 5 ORGANISM_TAXID: 562;
COMPND 10 EC: 3.6.1.-;
KEYWDS DNA HELICASE, HYDROLASE/DNA COMPLEX
SCOP: none