##########
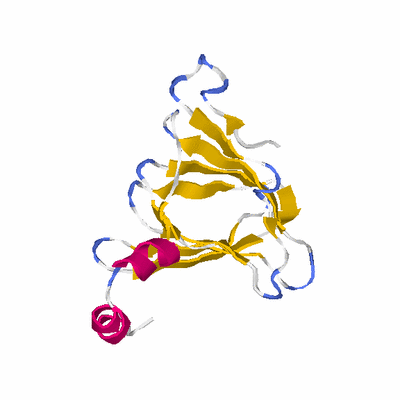
# PROTEIN: Q2246_545_419.5wLII_11277_198 231 419 # Category_C1
# Length: 231
# Weight: 25921.76
# Similarity: UniProt_B6AM57, BLAST_B6AM57, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 50.22 M_32 2uya_A 383 1 4e-04 17.000 116:126 (67-192:67-182)
# Evl 67.97 M_00 1y9q_A 192 1 9e-26 9.000 157:194 (3-183:12-182)
# Sid 20.35 M_13 2cvh_A 220 4 0.028 32.000 47:50 (180-229:36-82)
# Cov 78.79 M_02 3bdn_A 236 2 2e-25 14.000 182:232 (4-223:15-208)
# LaL 30.74 M_30 2k9q_A 77 2 4e-05 21.000 71:71 (1-71:1-71)
# OvN 26.84 M_04 1utx_A 66 4 4e-15 14.000 62:64 (1-64:1-62)
# OvC 20.35 M_13 2cvh_A 220 4 0.028 32.000 47:50 (180-229:36-82)
PDB: 2uya PDBsum
HEADER LYASE 03-APR-07 2UYA
TITLE DEL162-163 MUTANT OF BACILLUS SUBTILIS OXALATE
TITLE 2 DECARBOXYLASE OXDC
SOURCE 2 ORGANISM_SCIENTIFIC: BACILLUS SUBTILIS;
SOURCE 3 ORGANISM_TAXID: 1423;
COMPND 5 EC: 4.1.1.2;
KEYWDS LYASE, CUPIN, FORMATE, OXALATE, MANGANESE, METAL BINDING
KEYWDS 2 PROTEIN, METAL-BINDING, DECARBOXYLASE, DEL162-163 MUTANT
SCOP: none
PDB: 1y9q PDBsum
HEADER TRANSCRIPTION REGULATOR 16-DEC-04 1Y9Q
TITLE CRYSTAL STRUCTURE OF HTH_3 FAMILY TRANSCRIPTIONAL REGULATOR
TITLE 2 FROM VIBRIO CHOLERAE
SOURCE 2 ORGANISM_SCIENTIFIC: VIBRIO CHOLERAE;
SOURCE 3 ORGANISM_TAXID: 666;
KEYWDS HTH_3 FAMILY, TRANSCRIPTIONAL REGULAATOR, STRUCUTRAL
KEYWDS 2 GENOMICS, PROTEIN STRUCTURE INITIATIVE, PSI, NEW YORK SGX
KEYWDS 3 RESEARCH CENTER FOR STRUCTURAL GENOMICS, NYSGXRC, DIMER,
KEYWDS 4 TWO-DOMAIN STRUCTURE, STRUCTURAL GENOMICS, TRANSCRIPTION
KEYWDS 5 REGULATOR
SCOP: a.35 lambda repressor-like DNA-binding domains
SCOP: a.35.1 lambda repressor-like DNA-binding domains
SCOP: a.35.1.8 Probable transcriptional regulator VC1968, N-terminal domain
SCOP: b.82 Double-stranded beta-helix
SCOP: b.82.1 RmlC-like cupins
SCOP: b.82.1.15 Probable transcriptional regulator VC1968, C-terminal domain
PDB: 2cvh PDBsum
HEADER DNA BINDING PROTEIN 03-JUN-05 2CVH
TITLE CRYSTAL STRUCTURE OF THE RADB RECOMBINASE
SOURCE 2 ORGANISM_SCIENTIFIC: THERMOCOCCUS KODAKARENSIS;
SOURCE 3 ORGANISM_TAXID: 311400;
KEYWDS RADB, FILAMENT FORMATION, HOMOLOGOUS RECOMBINATION, ATPASE
KEYWDS 2 DOMAIN, HYPERTHERMOPHILE, DNA BINDING PROTEIN
SCOP: none
PDB: 3bdn PDBsum
HEADER TRANSCRIPTION/DNA 15-NOV-07 3BDN
TITLE CRYSTAL STRUCTURE OF THE LAMBDA REPRESSOR
SOURCE 2 ORGANISM_SCIENTIFIC: ENTEROBACTERIA PHAGE LAMBDA;
SOURCE 3 ORGANISM_TAXID: 10710;
KEYWDS LAMBDA, REPRESSOR, ALLOSTERY, COOPERATIVITY, DNA BINDING,
KEYWDS 2 TRANSCRIPTION/DNA COMPLEX
SCOP: none
PDB: 2k9q PDBsum
HEADER STRUCTURAL GENOMICS, UNKNOWN FUNCTION 23-OCT-08 2K9Q
TITLE SOLUTION NMR STRUCTURE OF HTH_XRE FAMILY TRANSCRIPTIONAL
TITLE 2 REGULATOR BT_P548217 FROM BACTEROIDES THETAIOTAOMICRON.
TITLE 3 NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET BTR244.
SOURCE 2 ORGANISM_SCIENTIFIC: BACTEROIDES THETAIOTAOMICRON;
SOURCE 3 ORGANISM_TAXID: 818;
KEYWDS ALL HELIX, HELIX-TURN-HELIX, PLASMID, STRUCTURAL GENOMICS,
KEYWDS 2 PSI-2, PROTEIN STRUCTURE INITIATIVE, NORTHEAST STRUCTURAL
KEYWDS 3 GENOMICS CONSORTIUM, NESG, UNKNOWN FUNCTION
SCOP: none
PDB: 1utx PDBsum
HEADER DNA-BINDING PROTEIN 12-DEC-03 1UTX
TITLE REGULATION OF CYTOLYSIN EXPRESSION BY ENTEROCOCCUS
TITLE 2 FAECALIS: ROLE OF CYLR2
SOURCE 2 ORGANISM_SCIENTIFIC: ENTEROCOCCUS FAECALIS;
SOURCE 3 ORGANISM_TAXID: 1351;
KEYWDS DNA-BINDING PROTEIN, TRANSCRIPTIONAL REPRESSOR, REGULATION
KEYWDS 2 OF CYTOLYSIN OPERON, HELIX-TURN-HELIX
SCOP: a.35 lambda repressor-like DNA-binding domains
SCOP: a.35.1 lambda repressor-like DNA-binding domains
SCOP: a.35.1.3 SinR domain-like