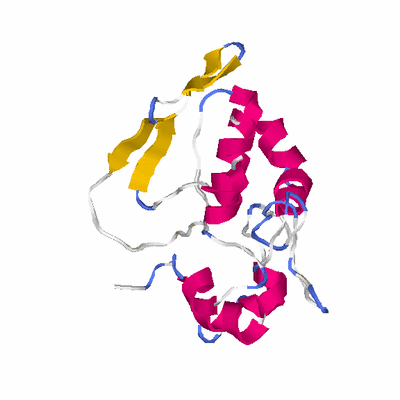
##########
# PROTEIN: Q2246_545_421.5wLII_11277_202 217 421 # Category_C1
# Length: 217
# Weight: 25249.65
# Similarity: UniProt_A3EPK0, BLAST_A3EPK0, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 64.06 M_00 1k6m_A 432 2 3e-38 20.000 139:161 (3-159:287-429)
# Evl 61.29 M_00 2odo_A 357 4 3e-42 14.000 133:163 (62-214:1-145)
# Sid 64.06 M_00 1k6m_A 432 2 3e-38 20.000 139:161 (3-159:287-429)
# Cov 69.59 M_00 1a0p_A 290 1 2e-30 14.000 151:188 (34-214:69-235)
# LaL 38.71 M_04 3c2w_C 505 8 0.051 13.000 84:88 (119-206:107-190)
# OvN 56.22 M_08 3d8h_B 267 2 8e-25 11.000 122:160 (2-135:103-260)
# OvC 63.13 M_02 3b8w_A 379 4 7e-41 10.000 137:168 (62-217:21-170)
PDB: 1k6m PDBsum
HEADER TRANSFERASE, HYDROLASE 16-OCT-01 1K6M
TITLE CRYSTAL STRUCTURE OF HUMAN LIVER 6-PHOSPHOFRUCTO-2-
TITLE 2 KINASE/FRUCTOSE-2,6-BISPHOSPHATASE
SOURCE 2 ORGANISM_SCIENTIFIC: HOMO SAPIENS;
SOURCE 3 ORGANISM_COMMON: HUMAN;
SOURCE 4 ORGANISM_TAXID: 9606;
COMPND 5 EC: 2.7.1.105, 3.1.3.46;
KEYWDS DOMAIN, TISSUE DIFFERENTIATION, ISOFORM, DOMAIN STABILITY,
KEYWDS 2 TRANSFERASE, HYDROLASE
SCOP: c.37 P-loop containing nucleoside triphosphate hydrolases
SCOP: c.37.1 P-loop containing nucleoside triphosphate hydrolases
SCOP: c.37.1.7 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase, kinase domain
SCOP: c.60 Phosphoglycerate mutase-like
SCOP: c.60.1 Phosphoglycerate mutase-like
SCOP: c.60.1.4 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase, phosphatase domain
PDB: 2odo PDBsum
HEADER ISOMERASE 25-DEC-06 2ODO
TITLE CRYSTAL STRUCTURE OF PSEUDOMONAS FLUORESCENS ALANINE
TITLE 2 RACEMASE
SOURCE 2 ORGANISM_SCIENTIFIC: PSEUDOMONAS FLUORESCENS;
SOURCE 3 ORGANISM_TAXID: 294;
COMPND 4 EC: 5.1.1.1;
KEYWDS ALANINE RACEMASE, PLP, ISOMERASE
SCOP: none
PDB: 1a0p PDBsum
HEADER DNA RECOMBINATION 05-DEC-97 1A0P
TITLE SITE-SPECIFIC RECOMBINASE, XERD
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
SOURCE 3 ORGANISM_TAXID: 562;
KEYWDS XERD, RECOMBINASE, DNA BINDING, DNA RECOMBINATION
SCOP: a.60 SAM domain-like
SCOP: a.60.9 lambda integrase-like, N-terminal domain
SCOP: a.60.9.1 lambda integrase-like, N-terminal domain
SCOP: d.163 DNA breaking-rejoining enzymes
SCOP: d.163.1 DNA breaking-rejoining enzymes
SCOP: d.163.1.1 Lambda integrase-like, catalytic core
PDB: 3c2w PDBsum
HEADER SIGNALING PROTEIN 25-JAN-08 3C2W
TITLE CRYSTAL STRUCTURE OF THE PHOTOSENSORY CORE DOMAIN OF P.
TITLE 2 AERUGINOSA BACTERIOPHYTOCHROME PABPHP IN THE PFR STATE
SOURCE 2 ORGANISM_SCIENTIFIC: PSEUDOMONAS AERUGINOSA;
SOURCE 3 ORGANISM_TAXID: 287;
COMPND 6 EC: 2.7.13.3;
KEYWDS KNOT STRUCTURE, CHROMOPHORE, KINASE, PHOSPHOPROTEIN,
KEYWDS 2 PHOTORECEPTOR PROTEIN, RECEPTOR, SENSORY TRANSDUCTION,
KEYWDS 3 TRANSFERASE, SIGNALING PROTEIN
SCOP: none
PDB: 3d8h PDBsum
HEADER ISOMERASE 23-MAY-08 3D8H
TITLE CRYSTAL STRUCTURE OF PHOSPHOGLYCERATE MUTASE FROM
TITLE 2 CRYPTOSPORIDIUM PARVUM, CGD7_4270
SOURCE 2 ORGANISM_SCIENTIFIC: CRYPTOSPORIDIUM PARVUM IOWA II;
SOURCE 3 ORGANISM_TAXID: 353152;
KEYWDS STRUCTURAL GENOMICS, MALARIA, CRYPTOSPORIDIUM, GLYCOLYSIS,
KEYWDS 2 ISOMERASE, STRUCTURAL GENOMICS CONSORTIUM, SGC
SCOP: none
PDB: 3b8w PDBsum
HEADER ISOMERASE 02-NOV-07 3B8W
TITLE CRYSTAL STRUCTURE OF ESCHERICHIA COLI ALAINE RACEMASE
TITLE 2 MUTANT E221P
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
SOURCE 3 ORGANISM_TAXID: 83333;
COMPND 4 EC: 5.1.1.1;
KEYWDS ALPHA/BETA BARREL, CELL SHAPE, CELL WALL
KEYWDS 2 BIOGENESIS/DEGRADATION, ISOMERASE, PEPTIDOGLYCAN SYNTHESIS,
KEYWDS 3 PYRIDOXAL PHOSPHATE
SCOP: none