##########
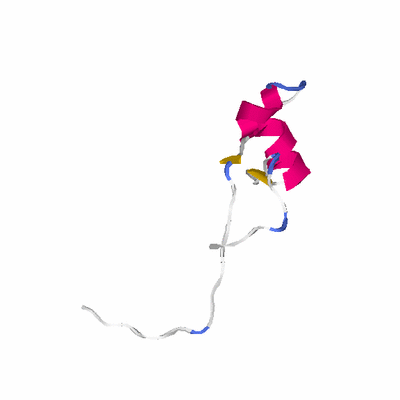
# PROTEIN: Q2246_545_427.5wLII_11277_221 90 427 # Category_C1
# Length: 90
# Weight: 10218.68
# Similarity: UniProt_B6AM78, BLAST_B6AM78, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 56.67 M_00 1hnj_A 317 11 4e-18 19.000 51:52 (35-85:2-53)
# Evl 56.67 M_00 1hnj_A 317 11 4e-18 19.000 51:52 (35-85:2-53)
# Sid 53.33 M_12 z0un 334 0 7e-14 26.000 48:49 (36-83:10-58)
# Cov 96.67 M_03 1jch_A 551 2 0.067 4.000 87:92 (2-88:280-371)
# LaL 41.11 M_03 3ezu_A 342 1 0.084 8.000 37:37 (36-72:176-212)
# OvN 46.67 M_00 2pr5_A 132 4 5e-05 16.000 42:49 (2-48:83-126)
# OvC 57.78 M_04 1ub7_A 322 4 8e-17 20.000 52:55 (36-89:2-54)
PDB: 1hnj PDBsum
HEADER TRANSFERASE 07-DEC-00 1HNJ
TITLE CRYSTAL STRUCTURE OF BETA-KETOACYL-ACP SYNTHASE III +
TITLE 2 MALONYL-COA
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
SOURCE 3 ORGANISM_TAXID: 562;
COMPND 6 EC: 2.3.1.41;
KEYWDS FABH, TRANSFERASE
SCOP: c.95 Thiolase-like
SCOP: c.95.1 Thiolase-like
SCOP: c.95.1.2 Chalcone synthase-like
PDB: 1hzp PDBsum
HEADER TRANSFERASE 25-JAN-01 1HZP
TITLE CRYSTAL STRUCTURE OF THE MYOBACTERIUM TUBERCULOSIS BETA-
TITLE 2 KETOACYL-ACYL CARRIER PROTEIN SYNTHASE III
SOURCE 2 ORGANISM_SCIENTIFIC: MYCOBACTERIUM TUBERCULOSIS;
SOURCE 3 ORGANISM_TAXID: 1773;
COMPND 5 EC: 2.3.1.41;
KEYWDS FATTY ACID BIOSYNTHESIS, MYOBACTERIUM TUBERCULOSIS,
KEYWDS 2 STRUCTURAL BASIS FOR SUBSTRATE SPECIFICITY, STRUCTURAL
KEYWDS 3 GENOMICS, PSI, PROTEIN STRUCTURE INITIATIVE, TB STRUCTURAL
KEYWDS 4 GENOMICS CONSORTIUM, TBSGC, TRANSFERASE
SCOP: c.95 Thiolase-like
SCOP: c.95.1 Thiolase-like
SCOP: c.95.1.2 Chalcone synthase-like
PDB: 1jch PDBsum
HEADER RIBOSOME INHIBITOR, HYDROLASE 09-JUN-01 1JCH
TITLE CRYSTAL STRUCTURE OF COLICIN E3 IN COMPLEX WITH ITS
TITLE 2 IMMUNITY PROTEIN
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI STR. K12 SUBSTR.
SOURCE 4 ORGANISM_TAXID: 316407;
SOURCE 10 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI STR. K12 SUBSTR.
COMPND 5 EC: 3.1.21.-;
KEYWDS TRANSLOCATION DOMAIN IS A BETA-JELLYROLL, THE RECEPTOR-
KEYWDS 2 BINDING DOMAIN IS A COILED COIL, THE RNASE DOMAIN IS A SIX-
KEYWDS 3 STRANDED ANTIPARALLEL BETA-SHEET. THE IMMUNITY PROTEIN IS A
KEYWDS 4 FOUR-STRANDED ANTIPARALLEL BETA SHEET FLANKED BY 3 HELICES
KEYWDS 5 ON ONE SIDE OF THE SHEET, RIBOSOME INHIBITOR, HYDROLASE
SCOP: b.101 Ribonuclease domain of colicin E3
SCOP: b.101.1 Ribonuclease domain of colicin E3
SCOP: b.101.1.1 Ribonuclease domain of colicin E3
SCOP: b.110 Cloacin translocation domain
SCOP: b.110.1 Cloacin translocation domain
SCOP: b.110.1.1 Cloacin translocation domain
SCOP: d.26 FKBP-like
SCOP: d.26.2 Colicin E3 immunity protein
SCOP: d.26.2.1 Colicin E3 immunity protein
SCOP: h.4 Antiparallel coiled-coil
SCOP: h.4.9 Colicin E3 receptor domain
SCOP: h.4.9.1 Colicin E3 receptor domain
PDB: 3ezu PDBsum
HEADER SIGNALING PROTEIN 23-OCT-08 3EZU
TITLE CRYSTAL STRUCTURE OF MULTIDOMAIN PROTEIN OF UNKNOWN
TITLE 2 FUNCTION WITH GGDEF-DOMAIN (NP_951600.1) FROM GEOBACTER
TITLE 3 SULFURREDUCENS AT 1.95 A RESOLUTION
SOURCE 2 ORGANISM_SCIENTIFIC: GEOBACTER SULFURREDUCENS;
SOURCE 3 ORGANISM_TAXID: 35554;
KEYWDS NP_951600.1, MULTIDOMAIN PROTEIN OF UNKNOWN FUNCTION WITH
KEYWDS 2 GGDEF-DOMAIN, STRUCTURAL GENOMICS, JOINT CENTER FOR
KEYWDS 3 STRUCTURAL GENOMICS, JCSG, PROTEIN STRUCTURE INITIATIVE,
KEYWDS 4 PSI-2, GGDEF DOMAIN, UNKNOWN FUNCTION, SIGNALING PROTEIN
SCOP: none
PDB: 2pr5 PDBsum
HEADER FLAVOPROTEIN, SIGNALING PROTEIN 03-MAY-07 2PR5
TITLE STRUCTURAL BASIS FOR LIGHT-DEPENDENT SIGNALING IN THE
TITLE 2 DIMERIC LOV PHOTOSENSOR YTVA (DARK STRUCTURE)
SOURCE 2 ORGANISM_SCIENTIFIC: BACILLUS SUBTILIS;
SOURCE 3 ORGANISM_TAXID: 1423;
KEYWDS LIGHT-OXYGEN-VOLTAGE, LOV, PER-ARNT-SIM, PAS, FLAVOPROTEIN,
KEYWDS 2 SIGNALING PROTEIN
SCOP: none
PDB: 1ub7 PDBsum
HEADER TRANSFERASE 31-MAR-03 1UB7
TITLE THE CRYSTAL ANALYSIS OF BETA-KEROACYL-[ACYL CARRIER
TITLE 2 PROTEIN] SYNTHASE III (FABH)FROM THERMUS THERMOPHILUS.
SOURCE 2 ORGANISM_SCIENTIFIC: THERMUS THERMOPHILUS HB8;
SOURCE 3 ORGANISM_TAXID: 300852;
COMPND 5 EC: 2.3.1.41;
KEYWDS FATTY ACID SYNTHESIS, BETA-KETOACYL-ACP SYNTHASE III, FABH,
KEYWDS 2 RIKEN STRUCTURAL GENOMICS/PROTEOMICS INITIATIVE, RSGI,
KEYWDS 3 STRUCTURAL GENOMICS, TRANSFERASE
SCOP: c.95 Thiolase-like
SCOP: c.95.1 Thiolase-like
SCOP: c.95.1.2 Chalcone synthase-like