##########
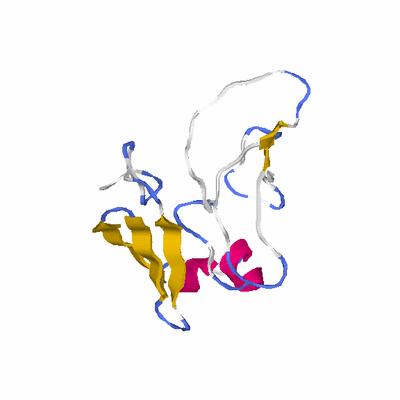
# PROTEIN: Q2246_545_460.5wLII_11346_17 146 460 # Category_C1
# Length: 146
# Weight: 17184.53
# Similarity: UniProt_B6AKY5, BLAST_B6AKY5, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 69.86 M_01 1v7w_A 807 3 5e-34 15.000 102:120 (31-139:102-214)
# Evl 77.40 M_00 2cqs_A 842 4 2e-34 11.000 113:138 (17-139:108-235)
# Sid 25.34 M_07 2p24_A 236 1 0.010 16.000 37:67 (23-87:5-71)
# Cov 93.84 M_00 1w66_A 232 1 1e-31 10.000 137:150 (1-143:14-157)
# LaL 58.90 M_06 1xdp_A 687 4 0.044 8.000 86:87 (43-129:504-589)
# OvN 93.84 M_00 1w66_A 232 1 1e-31 10.000 137:150 (1-143:14-157)
# OvC 68.49 M_01 1f0i_A 504 1 5e-06 11.000 100:115 (47-146:60-174)
PDB: 1v7w PDBsum
HEADER TRANSFERASE 24-DEC-03 1V7W
TITLE CRYSTAL STRUCTURE OF VIBRIO PROTEOLYTICUS CHITOBIOSE
TITLE 2 PHOSPHORYLASE IN COMPLEX WITH GLCNAC
SOURCE 2 ORGANISM_SCIENTIFIC: VIBRIO PROTEOLYTICUS;
SOURCE 3 ORGANISM_TAXID: 671;
COMPND 4 EC: 2.4.1.-;
KEYWDS BETA-SANDWICH, (ALPHA/ALPHA)6 BARREL, TRANSFERASE
SCOP: a.102 alpha/alpha toroid
SCOP: a.102.1 Six-hairpin glycosidases
SCOP: a.102.1.4 Glycosyltransferase family 36 C-terminal domain
SCOP: b.30 Supersandwich
SCOP: b.30.5 Galactose mutarotase-like
SCOP: b.30.5.3 Glycosyltransferase family 36 N-terminal domain
PDB: 2cqs PDBsum
HEADER TRANSFERASE 20-MAY-05 2CQS
TITLE CRYSTAL STRUCTURE OF CELLVIBRIO GILVUS CELLOBIOSE
TITLE 2 PHOSPHORYLASE CRYSTALLIZED FROM AMMONIUM SULFATE
SOURCE 2 ORGANISM_SCIENTIFIC: CELLVIBRIO GILVUS;
SOURCE 3 ORGANISM_TAXID: 11;
COMPND 4 EC: 2.4.1.20;
KEYWDS BETA-SANDWICH, (ALPHA/ALPHA)6 BARREL, TRANSFERASE
SCOP: none
PDB: 2p24 PDBsum
HEADER IMMUNE SYSTEM 06-MAR-07 2P24
TITLE I-AU/MBP125-135
SOURCE 2 ORGANISM_SCIENTIFIC: MUS MUSCULUS;
SOURCE 3 ORGANISM_COMMON: HOUSE MOUSE;
SOURCE 4 ORGANISM_TAXID: 10090;
KEYWDS MHC, I-AU, IMMUNOGLOBULIN FOLD, MBP, IMMUNE SYSTEM
SCOP: none
PDB: 1w66 PDBsum
HEADER TRANSFERASE 13-AUG-04 1W66
TITLE STRUCTURE OF A LIPOATE-PROTEIN LIGASE B FROM MYCOBACTERIUM
TITLE 2 TUBERCULOSIS
SOURCE 2 ORGANISM_SCIENTIFIC: MYCOBACTERIUM TUBERCULOSIS;
SOURCE 3 ORGANISM_TAXID: 83332;
COMPND 6 EC: 2.3.1.-;
KEYWDS LIPOATE-PROTEIN LIGASE B, LIPOYLTRANSFERASE, LIPOIC ACID,
KEYWDS 2 ACYLTRANSFERASE, TRANSFERASE, MYCOBACTERIUM TUBERCULOSIS
KEYWDS 3 STRUCTURAL PROTEOMICS PROJECT, XMTB, STRUCTURAL GENOMICS
SCOP: d.104 Class II aaRS and biotin synthetases
SCOP: d.104.1 Class II aaRS and biotin synthetases
SCOP: d.104.1.3 LplA-like
PDB: 1xdp PDBsum
HEADER TRANSFERASE 07-SEP-04 1XDP
TITLE CRYSTAL STRUCTURE OF THE E.COLI POLYPHOSPHATE KINASE IN
TITLE 2 COMPLEX WITH AMPPNP
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
SOURCE 3 ORGANISM_TAXID: 562;
COMPND 6 EC: 2.7.4.1;
KEYWDS E.COLI POLYPHOSPHATE KINASE, PPK, PPK COMPLEX WITH AMPPNP,
KEYWDS 2 AMPPNP, TRANSFERASE
SCOP: a.7 Spectrin repeat-like
SCOP: a.7.15 PPK N-terminal domain-like
SCOP: a.7.15.1 PPK N-terminal domain-like
SCOP: d.322 PHP14-like
SCOP: d.322.1 PHP14-like
SCOP: d.322.1.2 PPK middle domain-like
SCOP: d.136 Phospholipase D/nuclease
SCOP: d.136.1 Phospholipase D/nuclease
SCOP: d.136.1.4 Polyphosphate kinase C-terminal domain
PDB: 1f0i PDBsum
HEADER HYDROLASE 16-MAY-00 1F0I
TITLE THE FIRST CRYSTAL STRUCTURE OF A PHOSPHOLIPASE D
SOURCE 2 ORGANISM_SCIENTIFIC: STREPTOMYCES SP. PMF;
SOURCE 3 ORGANISM_TAXID: 172564;
COMPND 4 EC: 3.1.4.4
KEYWDS PHOSPHOLIPASE D, ALPHA-BETA-ALPHA-BETA-ALPHA STRUCTURE,
KEYWDS 2 HYDROLASE
SCOP: d.136 Phospholipase D/nuclease
SCOP: d.136.1 Phospholipase D/nuclease
SCOP: d.136.1.2 Phospholipase D