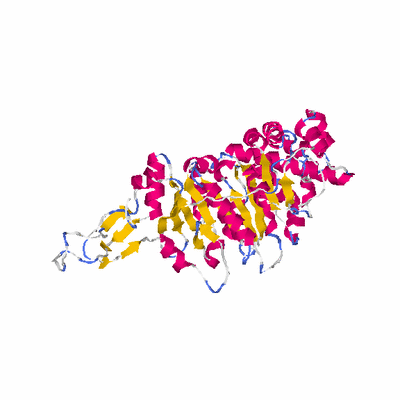
##########
# PROTEIN: Q2246_545_509.5wLII_11364_59 672 509 # Category_C1
# Length: 672
# Weight: 74793.14
# Similarity: UniProt_A3EUE4, BLAST_A3EUE4, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 61.01 M_00 1e9r_B 437 30 4e-76 28.000 410:443 (30-467:7-433)
# Evl 48.21 M_00 2ius_D 512 6 2e-85 14.000 324:427 (8-428:95-508)
# Sid 3.57 M_27 2pt7_A 330 6 0.031 45.000 24:24 (81-104:172-195)
# Cov 61.16 M_00 1e9r_B 437 30 2e-62 27.000 411:442 (30-467:7-433)
# LaL 9.23 M_07 1sxj_B 323 1 0.001 17.000 62:62 (51-112:13-74)
# OvN 16.07 M_02 2iuu_A 491 6 7e-05 16.000 108:122 (2-120:145-255)
# OvC 45.98 M_00 2ius_D 512 6 3e-79 12.000 309:504 (8-490:95-466)
PDB: 1e9r PDBsum
HEADER COUPLING PROTEIN 26-OCT-00 1E9R
TITLE BACTERIAL CONJUGATIVE COUPLING PROTEIN TRWBDELTAN70.
TITLE 2 TRIGONAL FORM IN COMPLEX WITH SULPHATE.
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
SOURCE 3 ORGANISM_TAXID: 562;
KEYWDS COUPLING PROTEIN, BACTERIAL CONJUGATION, F1-ATPASE-LIKE
KEYWDS 2 QUATERNARY STRUCTURE, RING HELICASES
SCOP: c.37 P-loop containing nucleoside triphosphate hydrolases
SCOP: c.37.1 P-loop containing nucleoside triphosphate hydrolases
SCOP: c.37.1.11 RecA protein-like (ATPase-domain)
PDB: 2ius PDBsum
HEADER MEMBRANE PROTEIN 07-JUN-06 2IUS
TITLE E. COLI FTSK MOTOR DOMAIN
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
SOURCE 3 ORGANISM_TAXID: 562;
KEYWDS NUCLEOTIDE-BINDING, CHROMOSOME PARTITION, ATP-BINDING, DNA-
KEYWDS 2 BINDING, CELL DIVISION, TRANSMEMBRANE, INNER MEMBRANE,
KEYWDS 3 HEXAMERIC RING, DNA TRANSLOCATION, KOPS, MEMBRANE,
KEYWDS 4 DIVISOME, CELL CYCLE, AAA ATPASE, MEMBRANE PROTEIN
SCOP: none
PDB: 2pt7 PDBsum
HEADER HYDROLASE/PROTEIN BINDING 08-MAY-07 2PT7
TITLE CRYSTAL STRUCTURE OF CAG VIRB11 (HP0525) AND AN INHIBITORY
TITLE 2 PROTEIN (HP1451)
SOURCE 2 ORGANISM_SCIENTIFIC: HELICOBACTER PYLORI 26695;
SOURCE 3 ORGANISM_TAXID: 85962;
SOURCE 12 ORGANISM_SCIENTIFIC: HELICOBACTER PYLORI 26695;
KEYWDS ATPASE, PROTEIN-PROTEIN COMPLEX, TYPE IV SECRETION,
KEYWDS 2 HYDROLASE/PROTEIN BINDING COMPLEX
SCOP: none
PDB: 1sxj PDBsum
HEADER REPLICATION 30-MAR-04 1SXJ
TITLE CRYSTAL STRUCTURE OF THE EUKARYOTIC CLAMP LOADER
TITLE 2 (REPLICATION FACTOR C, RFC) BOUND TO THE DNA SLIDING CLAMP
TITLE 3 (PROLIFERATING CELL NUCLEAR ANTIGEN, PCNA)
SOURCE 2 ORGANISM_SCIENTIFIC: SACCHAROMYCES CEREVISIAE;
SOURCE 3 ORGANISM_COMMON: BAKER'S YEAST;
SOURCE 4 ORGANISM_TAXID: 4932;
KEYWDS CLAMP LOADER, PROCESSIVITY CLAMP, DNA SLIDING CLAMP, AAA+
KEYWDS 2 ATPASE, DNA POLYMERASE, DNA-BINDING PROTEIN, REPLICATION
SCOP: a.80 DNA polymerase III clamp loader subunits, C-terminal domain
SCOP: a.80.1 DNA polymerase III clamp loader subunits, C-terminal domain
SCOP: a.80.1.1 DNA polymerase III clamp loader subunits, C-terminal domain
SCOP: c.37 P-loop containing nucleoside triphosphate hydrolases
SCOP: c.37.1 P-loop containing nucleoside triphosphate hydrolases
SCOP: c.37.1.20 Extended AAA-ATPase domain
SCOP: d.131 DNA clamp
SCOP: d.131.1 DNA clamp
SCOP: d.131.1.2 DNA polymerase processivity factor
PDB: 2iuu PDBsum
HEADER MEMBRANE PROTEIN 07-JUN-06 2IUU
TITLE P. AERUGINOSA FTSK MOTOR DOMAIN, HEXAMER
SOURCE 2 ORGANISM_SCIENTIFIC: PSEUDOMONAS AERUGINOSA;
SOURCE 3 ORGANISM_TAXID: 287;
KEYWDS DNA TRANSLOCATION, NUCLEOTIDE-BINDING, ATP-BINDING, DNA-
KEYWDS 2 BINDING, CELL DIVISION, TRANSMEMBRANE, CHROMOSOME
KEYWDS 3 PARTITION, INNER MEMBRANE, HEXAMERIC RING, MEMBRANE
KEYWDS 4 PROTEIN, KOPS, MEMBRANE, DIVISOME, CELL CYCLE, AAA ATPASE
SCOP: none