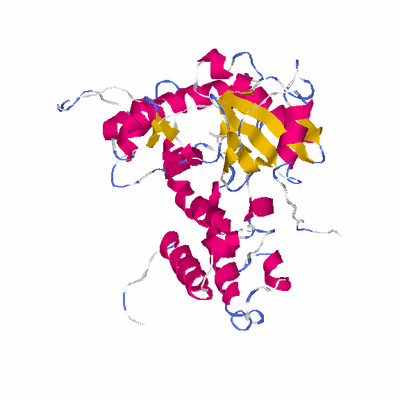
##########
# PROTEIN: Q2246_545_52.5wLII_10954_2 523 52 # Category_C1
# Length: 523
# Weight: 57313.20
# Similarity: UniProt_B6AQC8, BLAST_B6AQC8, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 56.02 M_01 z0d9 327 0 2e-28 18.000 293:364 (137-474:3-325)
# Evl 55.83 M_00 3dxp_A 359 1 8e-76 11.000 292:388 (132-490:8-348)
# Sid 4.59 M_37 2j51_A 325 2 0.074 37.000 24:24 (369-392:156-179)
# Cov 58.32 M_00 3dxp_A 359 1 8e-62 9.000 305:399 (127-497:7-357)
# LaL 9.94 M_35 3f69_B 311 2 0.070 15.000 52:52 (349-400:117-168)
# OvN 46.85 M_04 1nw1_A 429 2 5e-07 11.000 245:364 (107-429:8-330)
# OvC 24.09 M_37 2z7r_A 321 3 0.078 12.000 126:172 (354-522:135-275)
PDB: 1zyl PDBsum
HEADER STRUCTURAL GENOMICS, UNKNOWN FUNCTION 10-JUN-05 1ZYL
TITLE CRYSTAL STRUCTURE OF HYPOTHETICAL PROTEIN YIHE FROM
TITLE 2 ESCHERICHIA COLI
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
SOURCE 3 ORGANISM_TAXID: 562;
KEYWDS HYPOTHETICAL PROTEIN, PUTATIVE PROTEIN KINASE, STRUCTURAL
KEYWDS 2 GENOMICS, PSI, PROTEIN STRUCTURE INITIATIVE, MONTREAL-
KEYWDS 3 KINGSTON BACTERIAL STRUCTURAL GENOMICS INITIATIVE, BSGI,
KEYWDS 4 UNKNOWN FUNCTION
SCOP: none
PDB: 3dxp PDBsum
HEADER STRUCTURAL GENOMICS, UNKNOWN FUNCTION 24-JUL-08 3DXP
TITLE CRYSTAL STRUCTURE OF PUTATIVE ACYL-COA DEHYDROGENASE
TITLE 2 (YP_295230.1) FROM RALSTONIA EUTROPHA JMP134 AT 2.32 A
TITLE 3 RESOLUTION
SOURCE 2 ORGANISM_SCIENTIFIC: RALSTONIA EUTROPHA JMP134;
SOURCE 3 ORGANISM_COMMON: ALCALIGENES EUTROPHUS;
SOURCE 4 ORGANISM_TAXID: 264198;
KEYWDS YP_295230.1, PUTATIVE ACYL-COA DEHYDROGENASE, STRUCTURAL
KEYWDS 2 GENOMICS, JOINT CENTER FOR STRUCTURAL GENOMICS, JCSG,
KEYWDS 3 PROTEIN STRUCTURE INITIATIVE, PSI-2, TRANSFERASE, UNKNOWN
KEYWDS 4 FUNCTION
SCOP: none
PDB: 2j51 PDBsum
HEADER TRANSFERASE 08-SEP-06 2J51
TITLE CRYSTAL STRUCTURE OF HUMAN STE20-LIKE KINASE BOUND TO 5-
TITLE 2 AMINO-3-((4-(AMINOSULFONYL)PHENYL)AMINO)-N-(2,6-
TITLE 3 DIFLUOROPHENYL)-1H-1,2,4-TRIAZOLE-1-CARBOTHIOAMIDE
SOURCE 2 ORGANISM_SCIENTIFIC: HOMO SAPIENS;
SOURCE 3 ORGANISM_COMMON: HUMAN;
SOURCE 4 ORGANISM_TAXID: 9606;
COMPND 9 EC: 2.7.11.1;
KEYWDS TRANSFERASE, SERINE/THREONINE-PROTEIN KINASE, ATP-BINDING,
KEYWDS 2 PHOSPHORYLATION, MUSCLE DEVELOPMENT, KINASE, APOPTOSIS,
KEYWDS 3 COILED COIL, GERMINAL CENTRE KINASE, SERINE/THREONINE
KEYWDS 4 KINASE 2, NUCLEOTIDE-BINDING, ALTERNATIVE SPLICING
SCOP: none
PDB: 3f69 PDBsum
HEADER TRANSFERASE 05-NOV-08 3F69
TITLE CRYSTAL STRUCTURE OF THE MYCOBACTERIUM TUBERCULOSIS PKNB
TITLE 2 MUTANT KINASE DOMAIN IN COMPLEX WITH KT5720
SOURCE 2 ORGANISM_SCIENTIFIC: MYCOBACTERIUM TUBERCULOSIS;
SOURCE 3 ORGANISM_TAXID: 1773;
COMPND 5 EC: 2.7.11.1;
KEYWDS PROTEIN KINASE, PKNB, MYCOBACTERIUM TUBERCULOSIS, KT5720,
KEYWDS 2 STRUCTURAL GENOMICS, STRUCTUAL GENOMICS, COCRYSTALLIZATION
KEYWDS 3 OF PKNB KINASE DOMAIN AND INHIBITORS, TB STRUCTURAL
KEYWDS 4 GENOMICS CONSORTIUM, TBSGC, ATP-BINDING, KINASE, MAGNESIUM,
KEYWDS 5 METAL-BINDING, NUCLEOTIDE-BINDING, PHOSPHOPROTEIN,
KEYWDS 6 SERINE/THREONINE-PROTEIN KINASE, TRANSFERASE
SCOP: none
PDB: 1nw1 PDBsum
HEADER TRANSFERASE 05-FEB-03 1NW1
TITLE CRYSTAL STRUCTURE OF CHOLINE KINASE
SOURCE 2 ORGANISM_SCIENTIFIC: CAENORHABDITIS ELEGANS;
SOURCE 3 ORGANISM_TAXID: 6239;
COMPND 4 EC: 2.7.1.32;
KEYWDS CHOLINE KINASE, PHOSPHOLIPID SYNTHESIS, PROTEIN KINASE FOLD,
KEYWDS 2 TRANSFERASE
SCOP: d.144 Protein kinase-like (PK-like)
SCOP: d.144.1 Protein kinase-like (PK-like)
SCOP: d.144.1.8 Choline kinase
PDB: 2z7r PDBsum
HEADER TRANSFERASE 28-AUG-07 2Z7R
TITLE CRYSTAL STRUCTURE OF THE N-TERMINAL KINASE DOMAIN OF HUMAN
TITLE 2 RSK1 BOUND TO STAUROSPORINE
SOURCE 2 ORGANISM_SCIENTIFIC: HOMO SAPIENS;
SOURCE 3 ORGANISM_COMMON: HUMAN;
SOURCE 4 ORGANISM_TAXID: 9606;
COMPND 8 EC: 2.7.11.1;
KEYWDS PROTEIN KINASE, CANCER, KINASE INHIBITOR, ATP-BINDING,
KEYWDS 2 MAGNESIUM, METAL-BINDING, NUCLEOTIDE-BINDING,
KEYWDS 3 PHOSPHORYLATION, POLYMORPHISM, SERINE/THREONINE-PROTEIN
KEYWDS 4 KINASE, TRANSFERASE
SCOP: none