##########
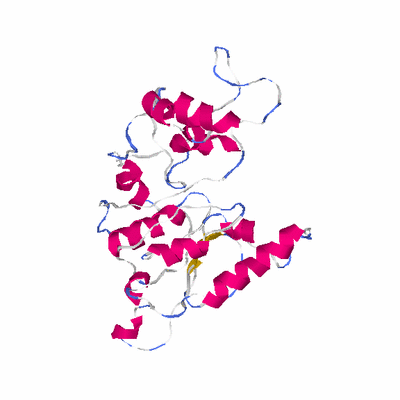
# PROTEIN: Q2246_545_525.5wLII_11390_11 300 525 # Category_C1
# Length: 300
# Weight: 34142.75
# Similarity: UniProt_B6ARI0, BLAST_B6ARI0, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 76.67 M_00 1ypw_A 806 10 5e-68 15.000 230:320 (1-276:437-746)
# Evl 72.67 M_00 2ce7_C 476 12 2e-74 17.000 218:281 (38-297:12-263)
# Sid 17.67 M_29 1sxj_C 340 1 1e-05 27.000 53:59 (33-90:16-69)
# Cov 80.33 M_09 2dhr_A 499 6 3e-58 12.000 241:300 (20-298:11-279)
# LaL 11.33 M_31 1g4a_E 443 8 1e-04 23.000 34:34 (67-100:50-83)
# OvN 76.33 M_02 1ypw_A 806 10 2e-68 16.000 229:292 (1-269:437-698)
# OvC 24.00 M_25 2dvw_B 83 3 6e-06 10.000 72:87 (213-299:2-76)
PDB: 1ypw PDBsum
HEADER TRANSPORT PROTEIN 31-JAN-05 1YPW
TITLE STRUCTURE OF P97/VCP IN COMPLEX IN ADP/AMP-PNP
SOURCE 2 ORGANISM_SCIENTIFIC: MUS MUSCULUS;
SOURCE 3 ORGANISM_COMMON: MOUSE;
KEYWDS AAA, P97/VCP, ERAD, CDC48
SCOP: none
PDB: 2ce7 PDBsum
HEADER CELL DIVISION PROTEIN 03-FEB-06 2CE7
TITLE EDTA TREATED
SOURCE 2 ORGANISM_SCIENTIFIC: THERMOTOGA MARITIMA;
SOURCE 3 ORGANISM_TAXID: 2336;
KEYWDS CELL DIVISION, METALLOPROTEASE, FTSH, CELL DIVISION PROTEIN
SCOP: a.269 FtsH protease domain-like
SCOP: a.269.1 FtsH protease domain-like
SCOP: a.269.1.1 FtsH protease domain-like
SCOP: c.37 P-loop containing nucleoside triphosphate hydrolases
SCOP: c.37.1 P-loop containing nucleoside triphosphate hydrolases
SCOP: c.37.1.20 Extended AAA-ATPase domain
PDB: 1sxj PDBsum
HEADER REPLICATION 30-MAR-04 1SXJ
TITLE CRYSTAL STRUCTURE OF THE EUKARYOTIC CLAMP LOADER
TITLE 2 (REPLICATION FACTOR C, RFC) BOUND TO THE DNA SLIDING CLAMP
TITLE 3 (PROLIFERATING CELL NUCLEAR ANTIGEN, PCNA)
SOURCE 2 ORGANISM_SCIENTIFIC: SACCHAROMYCES CEREVISIAE;
SOURCE 3 ORGANISM_COMMON: BAKER'S YEAST;
SOURCE 4 ORGANISM_TAXID: 4932;
KEYWDS CLAMP LOADER, PROCESSIVITY CLAMP, DNA SLIDING CLAMP, AAA+
KEYWDS 2 ATPASE, DNA POLYMERASE, DNA-BINDING PROTEIN, REPLICATION
SCOP: a.80 DNA polymerase III clamp loader subunits, C-terminal domain
SCOP: a.80.1 DNA polymerase III clamp loader subunits, C-terminal domain
SCOP: a.80.1.1 DNA polymerase III clamp loader subunits, C-terminal domain
SCOP: c.37 P-loop containing nucleoside triphosphate hydrolases
SCOP: c.37.1 P-loop containing nucleoside triphosphate hydrolases
SCOP: c.37.1.20 Extended AAA-ATPase domain
SCOP: d.131 DNA clamp
SCOP: d.131.1 DNA clamp
SCOP: d.131.1.2 DNA polymerase processivity factor
PDB: 2dhr PDBsum
HEADER HYDROLASE 24-MAR-06 2DHR
TITLE WHOLE CYTOSOLIC REGION OF ATP-DEPENDENT METALLOPROTEASE
TITLE 2 FTSH (G399L)
SOURCE 2 ORGANISM_SCIENTIFIC: THERMUS THERMOPHILUS;
SOURCE 3 ORGANISM_TAXID: 274;
KEYWDS AAA+ PROTEIN, HEXAMERIC ZN METALLOPROTEASE, HYDROLASE
SCOP: none
PDB: 1g4a PDBsum
HEADER CHAPERONE/HYDROLASE 26-OCT-00 1G4A
TITLE CRYSTAL STRUCTURES OF THE HSLVU PEPTIDASE-ATPASE COMPLEX
TITLE 2 REVEAL AN ATP-DEPENDENT PROTEOLYSIS MECHANISM
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
SOURCE 3 ORGANISM_TAXID: 562;
SOURCE 7 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
COMPND 11 EC: 3.4.99.-;
KEYWDS HSLVU, PEPTIDASE-ATPASE COMPLEX, CHAPERONE/HYDROLASE COMPLEX
SCOP: c.37 P-loop containing nucleoside triphosphate hydrolases
SCOP: c.37.1 P-loop containing nucleoside triphosphate hydrolases
SCOP: c.37.1.20 Extended AAA-ATPase domain
SCOP: d.153 Ntn hydrolase-like
SCOP: d.153.1 N-terminal nucleophile aminohydrolases (Ntn hydrolases)
SCOP: d.153.1.4 Proteasome subunits
PDB: 2dvw PDBsum
HEADER CELL CYCLE/PROTEIN-BINDING 01-AUG-06 2DVW
TITLE STRUCTURE OF THE ONCOPROTEIN GANKYRIN IN COMPLEX WITH S6
TITLE 2 ATPASE OF THE 26S PROTEASOME
SOURCE 2 ORGANISM_SCIENTIFIC: MUS MUSCULUS;
SOURCE 3 ORGANISM_COMMON: HOUSE MOUSE;
SOURCE 4 ORGANISM_TAXID: 10090;
KEYWDS ANKYRIN REPEATS, A-HELICAL DOMAIN, STRUCTURAL GENOMICS,
KEYWDS 2 NPPSFA, NATIONAL PROJECT ON PROTEIN STRUCTURAL AND
KEYWDS 3 FUNCTIONAL ANALYSES, RIKEN STRUCTURAL GENOMICS/PROTEOMICS
KEYWDS 4 INITIATIVE, RSGI, CELL CYCLE/PROTEIN-BINDING COMPLEX
SCOP: none