##########
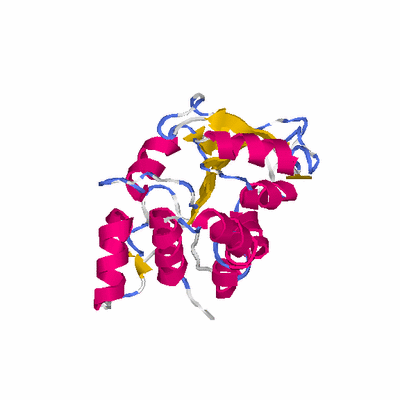
# PROTEIN: Q2246_545_532.5wLII_11390_53 388 532 # Category_C1
# Length: 388
# Weight: 43729.39
# Similarity: UniProt_B6ARM0, BLAST_B6ARM0, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 52.32 M_00 3f4k_A 257 0 7e-37 17.000 203:230 (68-292:30-237)
# Evl 69.59 M_00 1kph_B 287 6 1e-48 13.000 270:317 (35-341:8-287)
# Sid 31.19 M_36 z0oi 282 0 3e-12 22.000 121:150 (76-224:81-206)
# Cov 70.62 M_00 1kph_B 287 6 3e-41 13.000 274:313 (35-341:8-287)
# LaL 23.71 M_23 z0q6 173 0 4e-14 13.000 92:99 (139-237:3-95)
# OvN 57.73 M_01 1t43_A 277 1 3e-37 14.000 224:251 (2-244:19-250)
# OvC 69.59 M_00 1kph_B 287 6 1e-48 13.000 270:317 (35-341:8-287)
PDB: 3f4k PDBsum
HEADER TRANSFERASE 31-OCT-08 3F4K
TITLE CRYSTAL STRUCTURE OF A PROBABLE METHYLTRANSFERASE FROM
TITLE 2 BACTEROIDES THETAIOTAOMICRON. NORTHEAST STRUCTURAL
TITLE 3 GENOMICS TARGET BTR309.
SOURCE 2 ORGANISM_SCIENTIFIC: BACTEROIDES THETAIOTAOMICRON;
SOURCE 3 ORGANISM_TAXID: 818;
KEYWDS METHYLTRANSFERASE, STRUCTURAL GENOMICS, PSI-2, PROTEIN
KEYWDS 2 STRUCTURE INITIATIVE, NORTHEAST STRUCTURAL GENOMICS
KEYWDS 3 CONSORTIUM, NESG, TRANSFERASE
SCOP: none
PDB: 1kph PDBsum
HEADER TRANSFERASE 30-DEC-01 1KPH
TITLE CRYSTAL STRUCTURE OF MYCOLIC ACID CYCLOPROPANE SYNTHASE
TITLE 2 CMAA1 COMPLEXED WITH SAH AND DDDMAB
SOURCE 2 ORGANISM_SCIENTIFIC: MYCOBACTERIUM TUBERCULOSIS;
SOURCE 3 ORGANISM_TAXID: 1773;
COMPND 6 EC: 2.1.1.79;
KEYWDS MIXED ALPHA BETA FOLD, STRUCTURAL GENOMICS, PSI, PROTEIN
KEYWDS 2 STRUCTURE INITIATIVE, TB STRUCTURAL GENOMICS CONSORTIUM,
KEYWDS 3 TBSGC, TRANSFERASE
SCOP: c.66 S-adenosyl-L-methionine-dependent methyltransferases
SCOP: c.66.1 S-adenosyl-L-methionine-dependent methyltransferases
SCOP: c.66.1.18 Mycolic acid cyclopropane synthase
PDB: 1p91 PDBsum
HEADER TRANSFERASE 08-MAY-03 1P91
TITLE CRYSTAL STRUCTURE OF RLMA(I) ENZYME: 23S RRNA N1-G745
TITLE 2 METHYLTRANSFERASE (NORTHEAST STRUCTURAL GENOMICS
TITLE 3 CONSORTIUM TARGET ER19)
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
SOURCE 3 ORGANISM_TAXID: 562;
COMPND 6 EC: 2.1.1.51;
KEYWDS RLMA, RRMA, METHYLTRANSFERASE, G745, ER19, 23S RRNA, NESG,
KEYWDS 2 STRUCTURAL GENOMICS, PSI, PROTEIN STRUCTURE INITIATIVE,
KEYWDS 3 NORTHEAST STRUCTURAL GENOMICS CONSORTIUM
SCOP: c.66 S-adenosyl-L-methionine-dependent methyltransferases
SCOP: c.66.1 S-adenosyl-L-methionine-dependent methyltransferases
SCOP: c.66.1.33 rRNA methyltransferase RlmA
PDB: 1xxl PDBsum
HEADER STRUCTURAL GENOMICS, UNKNOWN FUNCTION 05-NOV-04 1XXL
TITLE THE CRYSTAL STRUCTURE OF YCGJ PROTEIN FROM BACILLUS
TITLE 2 SUBITILIS AT 2.1 A RESOLUTION
SOURCE 2 ORGANISM_SCIENTIFIC: BACILLUS SUBTILIS;
SOURCE 3 ORGANISM_TAXID: 1423;
KEYWDS STRUCTURAL GENOMICS, PROTEIN STRUCTURE INITIATIVE, PSI, NEW
KEYWDS 2 YORK SGX RESEARCH CENTER FOR STRUCTURAL GENOMICS, NYSGXRC,
KEYWDS 3 PROBABLE METHYLTRANSFERASE, UNKNOWN FUNCTION
SCOP: c.66 S-adenosyl-L-methionine-dependent methyltransferases
SCOP: c.66.1 S-adenosyl-L-methionine-dependent methyltransferases
SCOP: c.66.1.41 UbiE/COQ5-like
PDB: 1t43 PDBsum
HEADER TRANSFERASE 28-APR-04 1T43
TITLE CRYSTAL STRUCTURE ANALYSIS OF E.COLI PROTEIN (N5)-GLUTAMINE
TITLE 2 METHYLTRANSFERASE (HEMK)
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
SOURCE 3 ORGANISM_TAXID: 562;
COMPND 6 EC: 2.1.1.-;
KEYWDS METHYLTRANSFERASE
SCOP: c.66 S-adenosyl-L-methionine-dependent methyltransferases
SCOP: c.66.1 S-adenosyl-L-methionine-dependent methyltransferases
SCOP: c.66.1.30 N5-glutamine methyltransferase, HemK