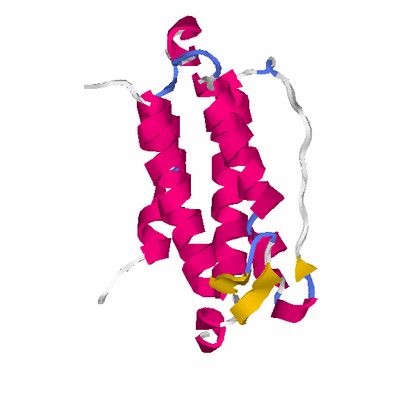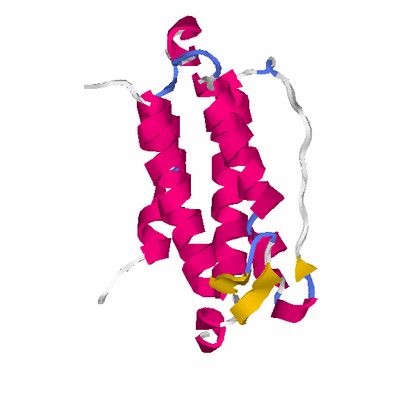##########
# PROTEIN: Q2246_545_538.5wLII_11390_77 175 538 # Category_B
# Length: 175
# Weight: 19612.54
# Similarity: UniProt_A3ESR7, BLAST_A3ESR7, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 77.14 M_08 2rd9_A 193 4 8e-06 20.000 135:145 (17-151:26-170)
# Evl 82.86 M_00 3di5_A 168 0 9e-26 18.000 145:169 (4-171:6-157)
# Sid 40.57 M_03 3dka_B 155 0 5e-04 23.000 71:78 (95-169:78-153)
# Cov 86.86 M_00 1rxq_B 178 4 3e-21 16.000 152:171 (3-159:12-177)
# LaL 42.29 M_07 2p1a_B 154 2 3e-06 14.000 74:75 (88-161:74-148)
# OvN 64.00 M_09 2yqy_A 169 2 0.034 15.000 112:164 (1-146:1-159)
# OvC 83.43 M_00 3di5_A 168 0 8e-20 17.000 146:174 (3-175:5-161)
PDB: 2rd9 PDBsum
HEADER STRUCTURAL GENOMICS, UNKNOWN FUNCTION 21-SEP-07 2RD9
TITLE CRYSTAL STRUCTURE OF YFIT-LIKE PUTATIVE METAL-DEPENDENT
TITLE 2 HYDROLASE (NP_241052.1) FROM BACILLUS HALODURANS AT 2.30 A
TITLE 3 RESOLUTION
SOURCE 2 ORGANISM_SCIENTIFIC: BACILLUS HALODURANS C-125;
SOURCE 3 ORGANISM_TAXID: 272558;
KEYWDS NP_241052.1, YFIT-LIKE PUTATIVE METAL-DEPENDENT HYDROLASE,
KEYWDS 2 STRUCTURAL GENOMICS, JOINT CENTER FOR STRUCTURAL GENOMICS,
KEYWDS 3 JCSG, PROTEIN STRUCTURE INITIATIVE, PSI-2, UNKNOWN FUNCTION
SCOP: none
PDB: 3di5 PDBsum
HEADER STRUCTURAL GENOMICS, UNKNOWN FUNCTION 19-JUN-08 3DI5
TITLE CRYSTAL STRUCTURE OF A DINB-LIKE PROTEIN (NP_980948.1) FROM
TITLE 2 BACILLUS CEREUS ATCC 10987 AT 2.01 A RESOLUTION
SOURCE 2 ORGANISM_SCIENTIFIC: BACILLUS CEREUS ATCC 10987;
SOURCE 3 ORGANISM_TAXID: 222523;
KEYWDS NP_980948.1, A DINB-LIKE PROTEIN, STRUCTURAL GENOMICS,
KEYWDS 2 JOINT CENTER FOR STRUCTURAL GENOMICS, JCSG, PROTEIN
KEYWDS 3 STRUCTURE INITIATIVE, PSI-2, DINB FAMILY, UNKNOWN FUNCTION
SCOP: none
PDB: 3dka PDBsum
HEADER STRUCTURAL GENOMICS, UNKNOWN FUNCTION 24-JUN-08 3DKA
TITLE CRYSTAL STRUCTURE OF A DINB-LIKE PROTEIN (NP_389123.1) FROM
TITLE 2 BACILLUS SUBTILIS AT 2.30 A RESOLUTION
SOURCE 2 ORGANISM_SCIENTIFIC: BACILLUS SUBTILIS;
SOURCE 3 ORGANISM_TAXID: 1423;
KEYWDS NP_389123.1, A DINB-LIKE PROTEIN, STRUCTURAL GENOMICS,
KEYWDS 2 JOINT CENTER FOR STRUCTURAL GENOMICS, JCSG, PROTEIN
KEYWDS 3 STRUCTURE INITIATIVE, PSI-2, PHOSPHOPROTEIN, DINB FAMILY,
KEYWDS 4 UNKNOWN FUNCTION
SCOP: none
PDB: 1rxq PDBsum
HEADER METAL-BINDING PROTEIN 18-DEC-03 1RXQ
TITLE YFIT FROM BACILLUS SUBTILIS IS A PROBABLE METAL-DEPENDENT
TITLE 2 HYDROLASE WITH AN UNUSUAL FOUR-HELIX BUNDLE TOPOLOGY
SOURCE 2 ORGANISM_SCIENTIFIC: BACILLUS SUBTILIS;
SOURCE 3 ORGANISM_TAXID: 1423;
KEYWDS NICKEL-BINDING, HYDROLASE, HELIX-BUNDLE, STRUCTURAL
KEYWDS 2 GENOMICS, PSI, PROTEIN STRUCTURE INITIATIVE, MIDWEST CENTER
KEYWDS 3 FOR STRUCTURAL GENOMICS, MCSG, METAL-BINDING PROTEIN
SCOP: a.213 DinB/YfiT-like putative metalloenzymes
SCOP: a.213.1 DinB/YfiT-like putative metalloenzymes
SCOP: a.213.1.1 YfiT-like putative metal-dependent hydrolases
PDB: 2p1a PDBsum
HEADER STRUCTURAL GENOMICS, UNKNOWN FUNCTION 02-MAR-07 2P1A
TITLE CRYSTAL STRUCTURE OF PUTATIVE DNA-BINDING PROTEIN FROM
TITLE 2 BACILLUS CEREUS ATCC 10987 AT 2.10 A RESOLUTION
SOURCE 2 ORGANISM_SCIENTIFIC: BACILLUS CEREUS ATCC 10987;
SOURCE 3 ORGANISM_TAXID: 222523;
KEYWDS NP_978475.1, HYPOTHETICAL PROTEIN, STRUCTURAL GENOMICS,
KEYWDS 2 JOINT CENTER FOR STRUCTURAL GENOMICS, JCSG, PROTEIN
KEYWDS 3 STRUCTURE INITIATIVE, PSI-2, UNKNOWN FUNCTION
SCOP: none
PDB: 2yqy PDBsum
HEADER STRUCTURAL GENOMICS, UNKNOWN FUNCTION 31-MAR-07 2YQY
TITLE CRYSTAL STRUCTURE OF TT2238, A FOUR-HELIX BUNDLE PROTEIN
SOURCE 2 ORGANISM_SCIENTIFIC: THERMUS THERMOPHILUS HB8;
SOURCE 3 ORGANISM_TAXID: 300852;
KEYWDS FOUR-HELIX-BUNDLE, NPPSFA, NATIONAL PROJECT ON PROTEIN
KEYWDS 2 STRUCTURAL AND FUNCTIONAL ANALYSES, STRUCTURAL GENOMICS,
KEYWDS 3 UNKNOWN FUNCTION
SCOP: none