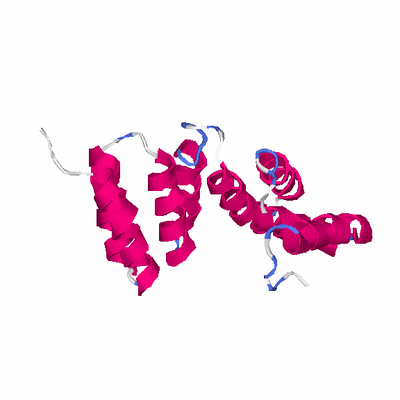
##########
# PROTEIN: Q2246_545_545.5wLII_11391_7 148 545 # Category_B
# Length: 148
# Weight: 17132.59
# Similarity: UniProt_B6AS38, BLAST_B6AS38, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 95.27 M_01 1w3b_A 388 2 1e-30 22.000 141:144 (4-147:235-375)
# Evl 88.51 M_00 2fo7_A 136 2 2e-33 18.000 131:133 (6-138:1-131)
# Sid 88.51 M_01 1w3b_A 388 2 1e-25 24.000 131:133 (4-136:235-365)
# Cov 95.27 M_01 1w3b_A 388 2 1e-30 22.000 141:144 (4-147:235-375)
# LaL 48.65 M_21 2dba_A 148 1 5e-11 20.000 72:72 (2-73:61-132)
# OvN 80.41 M_02 1na0_A 125 2 1e-29 20.000 119:124 (1-124:4-125)
# OvC 75.00 M_23 1xnf_B 275 2 5e-13 16.000 111:118 (31-148:35-145)
PDB: 1w3b PDBsum
HEADER TRANSFERASE 14-JUL-04 1W3B
TITLE THE SUPERHELICAL TPR DOMAIN OF O-LINKED GLCNAC TRANSFERASE
TITLE 2 REVEALS STRUCTURAL SIMILARITIES TO IMPORTIN ALPHA.
SOURCE 2 ORGANISM_SCIENTIFIC: HOMO SAPIENS;
SOURCE 3 ORGANISM_COMMON: HUMAN;
SOURCE 4 ORGANISM_TAXID: 9606;
COMPND 7 EC: 2.4.1.-;
KEYWDS OGT, GLCNAC, NUCLEOPORIN, O-LINKED GLYCOSYLATION, TPR
KEYWDS 2 REPEAT, PROTEIN BINDING, SIGNAL TRANSDUCTION, TRANSFERASE
SCOP: a.118 alpha-alpha superhelix
SCOP: a.118.8 TPR-like
SCOP: a.118.8.1 Tetratricopeptide repeat (TPR)
PDB: 2fo7 PDBsum
HEADER DE NOVO PROTEIN 12-JAN-06 2FO7
TITLE CRYSTAL STRUCTURE OF AN 8 REPEAT CONSENSUS TPR SUPERHELIX
TITLE 2 (TRIGONAL CRYSTAL FORM)
KEYWDS TETRATRICOPEPTIDE REPEAT, TPR, CONSENSUS PROTEIN,
KEYWDS 2 SUPERHELIX, DE NOVO PROTEIN
SCOP: k.38 TPR domain-based design
SCOP: k.38.1 TPR domain-based design
SCOP: k.38.1.1 TPR domain-based design
PDB: 2dba PDBsum
HEADER STRUCTURAL PROTEIN 15-DEC-05 2DBA
TITLE THE SOLUTION STRUCTURE OF THE TETRATRICO PEPTIDE REPEAT OF
TITLE 2 HUMAN SMOOTH MUSCLE CELL ASSOCIATED PROTEIN-1, ISOFORM 2
SOURCE 2 ORGANISM_SCIENTIFIC: HOMO SAPIENS;
SOURCE 3 ORGANISM_COMMON: HUMAN;
SOURCE 4 ORGANISM_TAXID: 9606;
KEYWDS TETRATRICOPEPTIDE REPEAT, SMOOTH MUSCLE CELL ASSOCIATED
KEYWDS 2 PROTEIN-1, STRUCTURAL GENOMICS, NPPSFA, NATIONAL PROJECT ON
KEYWDS 3 PROTEIN STRUCTURAL AND FUNCTIONAL ANALYSES, RIKEN
KEYWDS 4 STRUCTURAL GENOMICS/PROTEOMICS INITIATIVE, RSGI, STRUCTURAL
KEYWDS 5 PROTEIN
SCOP: none
PDB: 1na0 PDBsum
HEADER DE NOVO PROTEIN 26-NOV-02 1NA0
TITLE DESIGN OF STABLE ALPHA-HELICAL ARRAYS FROM AN IDEALIZED TPR
TITLE 2 MOTIF
SOURCE 2 ORGANISM_SCIENTIFIC: UNIDENTIFIED;
SOURCE 3 ORGANISM_TAXID: 32644;
KEYWDS DESIGN, TPR, DE NOVO PROTEIN
SCOP: k.38 TPR domain-based design
SCOP: k.38.1 TPR domain-based design
SCOP: k.38.1.1 TPR domain-based design
PDB: 1xnf PDBsum
HEADER STRUCTURAL GENOMICS, UNKOWN FUNCTION 04-OCT-04 1XNF
TITLE CRYSTAL STRUCTURE OF E.COLI TPR-PROTEIN NLPI
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
SOURCE 3 ORGANISM_TAXID: 562;
KEYWDS NLPI, TPR, TETRATRICOPEPTIDE, LIPOPROTEIN, STRUCTURAL
KEYWDS 2 GENOMICS, UNKOWN FUNCTION
SCOP: a.118 alpha-alpha superhelix
SCOP: a.118.8 TPR-like
SCOP: a.118.8.1 Tetratricopeptide repeat (TPR)