##########
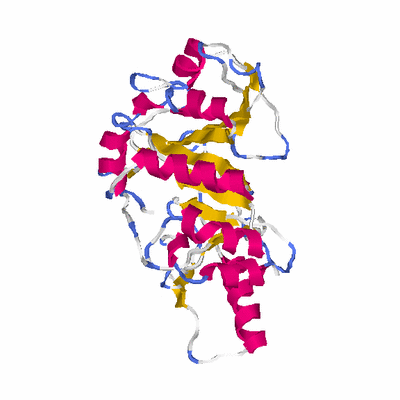
# PROTEIN: Q2246_545_79.5wLII_11067_27 462 79 # Category_C1
# Length: 462
# Weight: 51878.54
# Similarity: UniProt_B6APD8, BLAST_B6APD8, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 57.14 M_00 1tv8_A 340 6 2e-62 15.000 264:331 (69-353:6-315)
# Evl 32.25 M_00 1itw_A 741 8 2e-73 19.000 149:167 (302-456:496-656)
# Sid 5.84 M_17 1gq4_A 90 1 0.002 48.000 27:27 (7-33:30-56)
# Cov 60.61 M_00 1tv8_A 340 6 3e-48 13.000 280:339 (65-356:2-336)
# LaL 20.13 M_35 1n99_B 166 2 0.010 12.000 93:93 (1-93:25-117)
# OvN 14.29 M_06 2pzd_A 113 2 6e-13 26.000 66:67 (1-67:33-98)
# OvC 32.25 M_00 1itw_A 741 8 2e-73 19.000 149:167 (302-456:496-656)
PDB: 1tv8 PDBsum
HEADER LIGAND BINDING PROTEIN 28-JUN-04 1TV8
TITLE STRUCTURE OF MOAA IN COMPLEX WITH S-ADENOSYLMETHIONINE
SOURCE 2 ORGANISM_SCIENTIFIC: STAPHYLOCOCCUS AUREUS;
SOURCE 3 ORGANISM_TAXID: 1280;
KEYWDS TIM BARREL, LIGAND BINDING PROTEIN
SCOP: c.1 TIM beta/alpha-barrel
SCOP: c.1.28 Radical SAM enzymes
SCOP: c.1.28.3 MoCo biosynthesis proteins
PDB: 1itw PDBsum
HEADER OXIDOREDUCTASE 12-FEB-02 1ITW
TITLE CRYSTAL STRUCTURE OF THE MONOMERIC ISOCITRATE DEHYDROGENASE
TITLE 2 IN COMPLEX WITH ISOCITRATE AND MN
SOURCE 2 ORGANISM_SCIENTIFIC: AZOTOBACTER VINELANDII;
SOURCE 3 ORGANISM_TAXID: 354;
COMPND 4 EC: 1.1.1.42
KEYWDS GREECE KEY MOTIF, OXIDOREDUCTASE
SCOP: c.77 Isocitrate/Isopropylmalate dehydrogenase-like
SCOP: c.77.1 Isocitrate/Isopropylmalate dehydrogenase-like
SCOP: c.77.1.2 Monomeric isocitrate dehydrogenase
PDB: 1gq4 PDBsum
HEADER SIGNALING PROTEIN 19-NOV-01 1GQ4
TITLE STRUCTURAL DETERMINANTS OF THE NHERF INTERACTION WITH
TITLE 2 BETA2AR AND PDGFR
SOURCE 2 ORGANISM_SCIENTIFIC: HOMO SAPIENS;
SOURCE 3 ORGANISM_COMMON: HUMAN;
SOURCE 4 ORGANISM_TAXID: 9606;
KEYWDS SIGNALING PROTEIN, PDZ, BETA2-ADRENERGIC RECEPTOR, PDGFR,
KEYWDS 2 NHERF, CRYSTAL STRUCTURE, COMPLEX
SCOP: b.36 PDZ domain-like
SCOP: b.36.1 PDZ domain-like
SCOP: b.36.1.1 PDZ domain
PDB: 1n99 PDBsum
HEADER SIGNALING PROTEIN 22-NOV-02 1N99
TITLE CRYSTAL STRUCTURE OF THE PDZ TANDEM OF HUMAN SYNTENIN
SOURCE 2 ORGANISM_SCIENTIFIC: HOMO SAPIENS;
SOURCE 3 ORGANISM_COMMON: HUMAN;
SOURCE 4 ORGANISM_TAXID: 9606;
KEYWDS PDZ DOMAIN, SIGNALING PROTEIN
SCOP: b.36 PDZ domain-like
SCOP: b.36.1 PDZ domain-like
SCOP: b.36.1.1 PDZ domain
PDB: 2pzd PDBsum
HEADER HYDROLASE 17-MAY-07 2PZD
TITLE CRYSTAL STRUCTURE OF THE HTRA2/OMI PDZ DOMAIN BOUND TO A
TITLE 2 PHAGE-DERIVED LIGAND (WTMFWV)
SOURCE 2 ORGANISM_SCIENTIFIC: HOMO SAPIENS;
SOURCE 3 ORGANISM_COMMON: HUMAN;
SOURCE 4 ORGANISM_TAXID: 9606;
COMPND 7 EC: 3.4.21.108;
KEYWDS PDZ DOMAIN, SERINE PROTEASE, APOPTOSIS, MITOCHONDRIA,
KEYWDS 2 PEPTIDE-BINDING MODULE, HYDROLASE
SCOP: none