##########
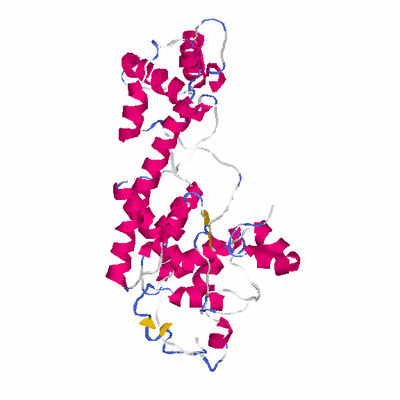
# PROTEIN: Q2246_545_286.5wLII_11238_21 381 286 # Category_C1
# Length: 381
# Weight: 42704.60
# Similarity: UniProt_B6AR02, BLAST_B6AR02, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 86.35 M_01 2nvu_A 536 1 e-111 15.000 329:379 (9-344:145-516)
# Evl 86.35 M_01 2nvu_A 536 1 e-111 15.000 329:379 (9-344:145-516)
# Sid 16.80 M_11 2it1_A 362 2 0.033 17.000 64:64 (235-298:5-68)
# Cov 86.35 M_01 2nvu_A 536 1 e-111 15.000 329:379 (9-344:145-516)
# LaL 16.80 M_11 2it1_A 362 2 0.033 17.000 64:64 (235-298:5-68)
# OvN 36.75 M_04 2hvd_A 152 9 9e-56 11.000 140:156 (1-146:1-151)
# OvC 86.35 M_01 2nvu_A 536 1 e-111 15.000 329:379 (9-344:145-516)
PDB: 2nvu PDBsum
HEADER PROTEIN TURNOVER, LIGASE 13-NOV-06 2NVU
TITLE STRUCTURE OF APPBP1-UBA3~NEDD8-NEDD8-MGATP-UBC12(C111A), A
TITLE 2 TRAPPED UBIQUITIN-LIKE PROTEIN ACTIVATION COMPLEX
SOURCE 2 ORGANISM_SCIENTIFIC: HOMO SAPIENS;
SOURCE 3 ORGANISM_COMMON: HUMAN;
SOURCE 4 ORGANISM_TAXID: 9606;
COMPND 15 EC: 6.3.2.-;
COMPND 26 EC: 6.3.2.-;
KEYWDS MULTIFUNCTION MACROMOLECULAR COMPLEX, UBIQUITIN, NEDD8, E1,
KEYWDS 2 E2, ATP, CONFORMATIONAL CHANGE, THIOESTER, SWITCH,
KEYWDS 3 ADENYLATION, PROTEIN TURNOVER, LIGASE
SCOP: c.111 Activating enzymes of the ubiquitin-like proteins
SCOP: c.111.1 Activating enzymes of the ubiquitin-like proteins
SCOP: c.111.1.2 Ubiquitin activating enzymes (UBA)
SCOP: c.94 Periplasmic binding protein-like II
SCOP: c.94.1 Periplasmic binding protein-like II
SCOP: c.94.1.1 Phosphate binding protein-like
SCOP: d.15 beta-Grasp (ubiquitin-like)
SCOP: d.15.1 Ubiquitin-like
SCOP: d.15.1.1 Ubiquitin-related
PDB: 2it1 PDBsum
HEADER TRANSPORT PROTEIN 18-OCT-06 2IT1
TITLE STRUCTURE OF PH0203 PROTEIN FROM PYROCOCCUS HORIKOSHII
SOURCE 2 ORGANISM_SCIENTIFIC: PYROCOCCUS HORIKOSHII;
SOURCE 3 ORGANISM_TAXID: 53953;
KEYWDS ATP BINDING PROTEIN, STRUCTURAL GENOMICS, NPPSFA, NATIONAL
KEYWDS 2 PROJECT ON PROTEIN STRUCTURAL AND FUNCTIONAL ANALYSES,
KEYWDS 3 RIKEN STRUCTURAL GENOMICS/PROTEOMICS INITIATIVE, RSGI,
KEYWDS 4 TRANSPORT PROTEIN
SCOP: none
PDB: 2hvd PDBsum
HEADER SIGNALING PROTEIN,TRANSFERASE 28-JUL-06 2HVD
TITLE HUMAN NUCLEOSIDE DIPHOSPHATE KINASE A COMPLEXED WITH ADP
SOURCE 2 ORGANISM_SCIENTIFIC: HOMO SAPIENS;
SOURCE 3 ORGANISM_COMMON: HUMAN;
SOURCE 4 ORGANISM_TAXID: 9606;
COMPND 7 EC: 2.7.4.6;
KEYWDS COMPLEX ADP, SIGNALING PROTEIN,TRANSFERASE
SCOP: d.58 Ferredoxin-like
SCOP: d.58.6 Nucleoside diphosphate kinase, NDK
SCOP: d.58.6.1 Nucleoside diphosphate kinase, NDK