##########
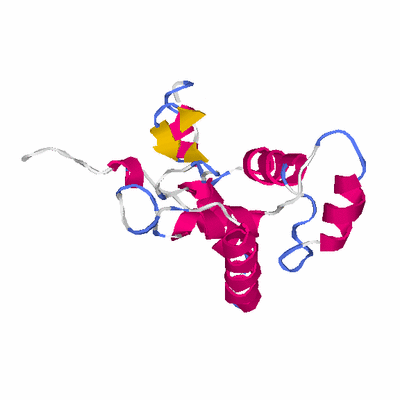
# PROTEIN: Q2246_545_464.5wLII_11346_28 193 464 # Category_C2
# Length: 193
# Weight: 21716.28
# Similarity: UniProt_B6AKZ5, BLAST_B6AKZ5, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 62.18 M_00 2oqo_A 200 2 2e-42 13.000 120:153 (48-193:6-140)
# Evl 62.18 M_00 2oqo_A 200 2 2e-42 13.000 120:153 (48-193:6-140)
# Sid 29.02 M_20 1knw_A 425 2 0.079 17.000 56:64 (117-180:94-149)
# Cov 70.98 M_01 3dwk_C 625 4 2e-35 12.000 137:165 (37-193:2-147)
# LaL 18.65 M_16 z0q6 173 0 0.095 2.000 36:36 (150-185:23-58)
# OvN 70.47 M_02 2olv_B 669 3 6e-35 12.000 136:170 (32-193:6-156)
# OvC 62.18 M_00 2oqo_A 200 2 2e-42 13.000 120:153 (48-193:6-140)
PDB: 2oqo PDBsum
HEADER TRANSFERASE 31-JAN-07 2OQO
TITLE CRYSTAL STRUCTURE OF A PEPTIDOGLYCAN GLYCOSYLTRANSFERASE
TITLE 2 FROM A CLASS A PBP: INSIGHT INTO BACTERIAL CELL WALL
TITLE 3 SYNTHESIS
SOURCE 2 ORGANISM_SCIENTIFIC: AQUIFEX AEOLICUS VF5;
SOURCE 3 ORGANISM_TAXID: 224324;
COMPND 5 EC: 2.4.2.-;
KEYWDS TRANSFERASE
SCOP: none
PDB: 1knw PDBsum
HEADER LYASE 19-DEC-01 1KNW
TITLE CRYSTAL STRUCTURE OF DIAMINOPIMELATE DECARBOXYLASE
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
SOURCE 3 ORGANISM_TAXID: 562;
COMPND 5 EC: 4.1.1.20;
KEYWDS PYRIDOXAL-PHOSPHATE, DECARBOXYLATION, DIAMINOPIMELATE,
KEYWDS 2 LYSINE, TIM-BARREL, LYASE
SCOP: b.49 Domain of alpha and beta subunits of F1 ATP synthase-like
SCOP: b.49.2 Alanine racemase C-terminal domain-like
SCOP: b.49.2.3 Eukaryotic ODC-like
SCOP: c.1 TIM beta/alpha-barrel
SCOP: c.1.6 PLP-binding barrel
SCOP: c.1.6.1 Alanine racemase-like, N-terminal domain
PDB: 3dwk PDBsum
HEADER TRANSFERASE 22-JUL-08 3DWK
TITLE IDENTIFICATION OF DYNAMIC STRUCTURAL MOTIFS INVOLVED IN
TITLE 2 PEPTIDOGLYCAN GLYCOSYLTRANSFER
SOURCE 2 ORGANISM_SCIENTIFIC: STAPHYLOCOCCUS AUREUS;
SOURCE 3 ORGANISM_TAXID: 93062;
KEYWDS LYSOZYME-FOLD TRANSPEPTIDASE FOLD PI-HELIX, CELL SHAPE,
KEYWDS 2 CELL WALL BIOGENESIS/DEGRADATION, MEMBRANE, PEPTIDOGLYCAN
KEYWDS 3 SYNTHESIS, TRANSFERASE
SCOP: none
PDB: 1xxl PDBsum
HEADER STRUCTURAL GENOMICS, UNKNOWN FUNCTION 05-NOV-04 1XXL
TITLE THE CRYSTAL STRUCTURE OF YCGJ PROTEIN FROM BACILLUS
TITLE 2 SUBITILIS AT 2.1 A RESOLUTION
SOURCE 2 ORGANISM_SCIENTIFIC: BACILLUS SUBTILIS;
SOURCE 3 ORGANISM_TAXID: 1423;
KEYWDS STRUCTURAL GENOMICS, PROTEIN STRUCTURE INITIATIVE, PSI, NEW
KEYWDS 2 YORK SGX RESEARCH CENTER FOR STRUCTURAL GENOMICS, NYSGXRC,
KEYWDS 3 PROBABLE METHYLTRANSFERASE, UNKNOWN FUNCTION
SCOP: c.66 S-adenosyl-L-methionine-dependent methyltransferases
SCOP: c.66.1 S-adenosyl-L-methionine-dependent methyltransferases
SCOP: c.66.1.41 UbiE/COQ5-like
PDB: 2olv PDBsum
HEADER TRANSFERASE 19-JAN-07 2OLV
TITLE STRUCTURAL INSIGHT INTO THE TRANSGLYCOSYLATION STEP OF
TITLE 2 BACTERIAL CELL WALL BIOSYNTHESIS : DONOR LIGAND COMPLEX
SOURCE 2 ORGANISM_SCIENTIFIC: STAPHYLOCOCCUS AUREUS;
SOURCE 3 ORGANISM_TAXID: 1280;
COMPND 4 EC: 2.3.2.-;
KEYWDS TRANSPEPTIDASE FOLD, GLYCOSYLTRANSFERASE FAMILY 51,
KEYWDS 2 LYSOZYME FOLD
SCOP: none